GenoPred Pipeline - Using 23andMe data
This document is a simplified set of instructions for the specific scenario where 23andMe data for an individual, and a set of score files on the PGS catalogue is being used as the input to the pipeline. It also provides a demonstration of using the genopred container.
Setting up
Download GenoPred container
Containers are available for both docker and singularity (see here)
singularity \
pull \
--arch \
amd64 \
/users/k1806347/oliverpainfel/Software/singularity/genopred_pipeline_latest.sif \
library://opain/genopred/genopred_pipeline:latestPrepare configuration
# Create directory to store configuration files, outputs and resources
workdir<-'/users/k1806347/oliverpainfel/test/genopred_23andme'
dir.create(paste0(workdir, '/config'), recursive = T)
dir.create(paste0(workdir, '/resources'), recursive = T)
dir.create(paste0(workdir, '/output'), recursive = T)
# Create config file
config<-c(
paste0('outdir: ', workdir, '/output'),
paste0('resdir: ', workdir, '/resources'),
paste0('config_file: ', workdir, '/config/config.yaml'),
paste0('target_list: ', workdir, '/config/target_list.txt'),
paste0('score_list: ', workdir, '/config/score_list.txt'),
'cores_target_pgs: 8',
'cores_impute_23andme: 8'
)
write.table(config, paste0(workdir, '/config/config.yaml'), col.names = F, row.names = F, quote = F)
# Create target_list
target_list<-data.frame(
name='Joe_Bloggs',
path='/users/k1806347/oliverpainfel/Software/MyGit/GenoPred/pipeline/test_data/target/23andMe_individual/Joe_Bloggs_genome_0123456789.zip',
type='23andMe',
indiv_report=TRUE
)
write.table(target_list, paste0(workdir, '/config/target_list.txt'), col.names=T, row.names=F, quote=F)
# Create score_list
score_list<-data.frame(
name='PGS002804',
path=NA,
label='Height'
)
score_list$label<-paste0("\"", score_list$label, "\"")
write.table(score_list, paste0(workdir, '/config/score_list.txt'), col.names=T, row.names=F, quote=F)Run pipeline
# Start container
singularity shell \
--bind /scratch/prj/oliverpainfel:/scratch/prj/oliverpainfel \
--writable-tmpfs \
/users/k1806347/oliverpainfel/Software/singularity/genopred_pipeline_latest.sif
# Activate GenoPred environment
source /opt/mambaforge/etc/profile.d/conda.sh
conda activate genopred
# Go to GenoPred/pipeline directory
cd /tools/GenoPred/pipeline/
# Do a dry run to check the scheduled steps are expected (there should not be any steps saying 'download', and it should not be necessary to build the conda environment)
snakemake -j8 --use-conda --configfile=/users/k1806347/oliverpainfel/test/genopred_23andme/config/config.yaml output_allComputational benchmark
Show code
bm_files_i<-list.files(path='/users/k1806347/oliverpainfel/test/genopred_23andme/output/reference/benchmarks', full.names = T)
bm_dat_all <- do.call(rbind, lapply(bm_files_i, function(file) {
tmp <- fread(file)
tmp$file <- basename(file)
return(tmp)
}))
# Create rule column
bm_dat_all$rule <- gsub('-.*','',bm_dat_all$file)
# Calculate total wall time
sum(bm_dat_all$s)/60/60 # 4.904476 hours
# Calculate wall time taken by rule
bm_dat_rule_time <- NULL
for (i in unique(bm_dat_all$rule)) {
bm_dat_rule_time <- rbind(
bm_dat_rule_time,
data.frame(
rule = i,
time = sum(bm_dat_all$s[bm_dat_all$rule == i])))
}
# Tidy results
bm_dat_rule_time <-
bm_dat_rule_time[!(bm_dat_rule_time$rule %in% c('ancestry_reporter', 'score_reporter.txt')), ]
bm_dat_rule_time$step<-gsub('_i$','', bm_dat_rule_time$rule)
bm_dat_rule_time$step<-gsub('prep_pgs_','', bm_dat_rule_time$step)
bm_dat_rule_time$step[bm_dat_rule_time$rule == 'format_target_i']<-'Target QC'
bm_dat_rule_time$step[bm_dat_rule_time$rule == 'impute_23andme_i']<-'Imputation'
bm_dat_rule_time$step[bm_dat_rule_time$rule == 'sumstat_prep_i']<-'GWAS QC'
bm_dat_rule_time$step[bm_dat_rule_time$rule == 'indiv_report_i']<-'Report Creation'
bm_dat_rule_time$step[bm_dat_rule_time$rule == 'ancestry_inference_i']<-'Ancestry Inference'
bm_dat_rule_time$step[bm_dat_rule_time$rule == 'target_pgs_i']<-'Target Scoring'
bm_dat_rule_time$step[bm_dat_rule_time$rule == 'prep_pgs_external_i']<-'Score file QC'
bm_dat_rule_time$step[bm_dat_rule_time$rule == 'download_pgs_external']<-'Download score file'
#######
# Create a pie chart
#######
data <- data.frame(
category = bm_dat_rule_time$step,
values = bm_dat_rule_time$time
)
data$category<-factor(data$category,
levels = c('Imputation',
'Target QC',
'Ancestry Inference',
'Download score file',
'Score file QC',
'Target Scoring',
'Report Creation'))
data<-data[order(data$category),]
data$perc<-data$values/sum(data$values)*100
data$cum_perc<-cumsum(data$perc)
for(i in 1:length(data$category)) {
data$start[i] <- ifelse(i == 1, 0, data$end[i - 1])
data$end[i] <- data$cum_perc[i]
data$label_position[i] <- data$cum_perc[i] - data$perc[i] / 2
}
data$time_clean<-NA
data$time_clean[data$values < 60] <-
paste0(round(data$values[data$values < 60], 1), ' sec')
data$time_clean[data$values > 60] <-
paste0(round(data$values[data$values > 60] / 60, 1), ' min')
data$time_clean[data$values > 3600] <-
paste0(round(data$values[data$values > 3600] / 60 / 60, 1), ' hr')
data$label<-paste0(data$category,
"\n(", data$time_clean, ', ',
round(data$perc,1),"%)")
library(ggplot2)
library(ggrepel)
png('~/oliverpainfel/Software/MyGit/GenoPred/docs/Images/Benchmark/23andme_benchmark.png', res=300, width=2500, height=1000, units = 'px')
set.seed(1)
ggplot(data,
aes(
xmin = start,
xmax = end,
ymin = 0,
ymax = 0.1,
fill = category
)) +
geom_rect(colour='black', size=0.1) +
geom_text_repel(
aes(
x = label_position,
y = 0.1,
label = label,
segment.square = T,
segment.inflect = T
),
force = 100,
nudge_y = 0.15,
hjust = 0.5,
segment.size = 0.3,
segment.curvature = -0.1,
segment.color='darkgrey',
box.padding=0.6,
) +
scale_x_continuous(breaks = seq(0, 100, by = 20)) +
coord_cartesian(clip = "off", xlim = c(-20, 120), ylim = c(0, 0.3)) +
labs(x='Time (%)') +
theme(
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.y = element_blank(),
axis.line.x = element_line(color = "black", size = 0.5),
panel.grid.major.y = element_blank(),
panel.grid.minor.y = element_blank(),
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
legend.position = "none",
plot.background = element_rect(fill = "white", color = NA),
panel.background = element_rect(fill = "white", color = NA)
)
dev.off()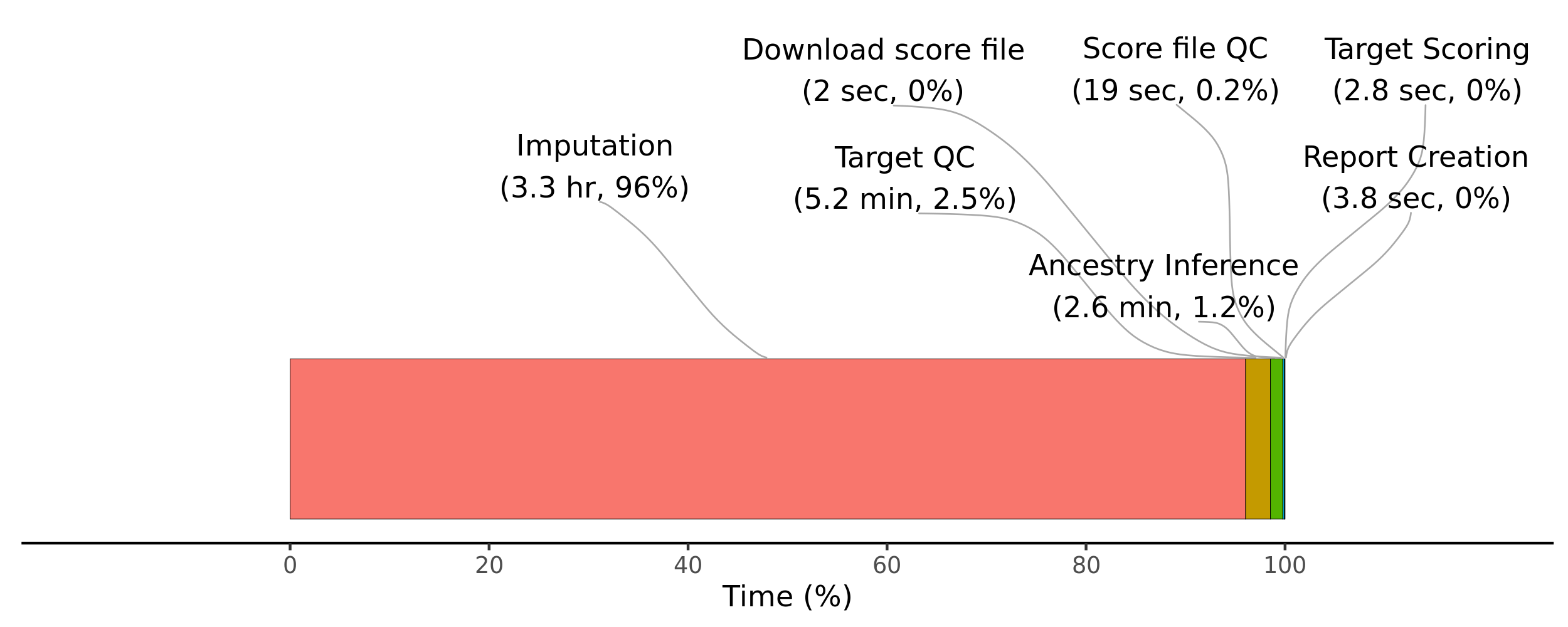
Time taken for each step

