Improving interpretability of polygenic scores using only summary statistics
This page describes a project investigatin approaches for converting polygenic scores into interpretable information.
Aims:
- Develop method for converting polygenic Z-scores into absolute estimates using summary statistics
- Develop figures to represent absolute risk
- Validate approach for estimating PRS R2/AUC from summary statistics
See here for a preprint describing this work.
1 Converting polygenic Z-scores to absolute estimates
To enable correct intepretation of a polygenic score, the variance explained by the polygenic score must be considered. Furthermore, for binary outcomes the population prevelance must be considered, and for continuous outcomes the population mean and SD must be considered. It is possible to convert relative genetic risk into absolute esimates of an outcome when observed data is available, as 23andMe do, by splitting participants into genetic risk quantiles, and then estimating the mean outcome within each quantile. However, observed data is often not available. Here, we use an alternative approach based on summary statistics only alone.
1.1 Describe conversion
1.1.1 Binary outcomes
To convert a polygenic Z-score into an absolute estimate of risk, we must know the predicitve utility of the polygenic score (AUC), and the prevelance of the outcome in the general population. Then it is possible to estimate the proportion of cases within each polygenic score quantile using bivariate-normal distribution.
Show code
# Thank you for Alex Gillet for her work developing this code.
ccprobs.f <- function(PRS_auc=0.641, prev=0.7463, n_quantile=20){
# Convert AUC into cohen's d
d <- sqrt(2)*qnorm(PRS_auc)
# Set mean difference between cases and control polygenic scores
mu_case <- d
mu_control <- 0
# Estimate mean and variance of polygenic scores across case and control
varPRS <- prev*(1+(d^2) - (d*prev)^2) + (1-prev)*(1 - (d*prev)^2)
E_PRS <- d*prev
# Estimate polygenic score quantiles
by_quant<-1/n_quantile
p_quant <- seq(by_quant, 1-by_quant, by=by_quant)
quant_vals_PRS <- rep(0, length(p_quant))
quant_f_solve <- function(x, prev, d, pq){prev*pnorm(x-d) + (1-prev)*pnorm(x) - pq}
for(i in 1:length(p_quant)){
quant_vals_PRS[i] <- unlist(uniroot(quant_f_solve, prev=prev, d=d, pq= p_quant[i], interval=c(-2.5, 2.5), extendInt = "yes", tol=6e-12)$root)
}
# Create a table for output
ul_qv_PRS <- matrix(0, ncol=2, nrow=n_quantile)
ul_qv_PRS[1,1] <- -Inf
ul_qv_PRS[2:n_quantile,1] <- quant_vals_PRS
ul_qv_PRS[1:(n_quantile-1),2] <- quant_vals_PRS
ul_qv_PRS[n_quantile,2] <- Inf
ul_qv_PRS<-cbind(ul_qv_PRS, (ul_qv_PRS[,1:2]-E_PRS)/sqrt(varPRS))
# Estimate case control proportion for each quantile
prob_quantile_case <- pnorm(ul_qv_PRS[,2], mean = mu_case) - pnorm(ul_qv_PRS[,1], mean = mu_case)
prob_quantile_control <- pnorm(ul_qv_PRS[,2], mean = mu_control) - pnorm(ul_qv_PRS[,1], mean = mu_control)
p_case_quantile <- (prob_quantile_case*prev)/by_quant
p_cont_quantile <- (prob_quantile_control*(1-prev))/by_quant
# Estimate OR comparing each quantile to bottom quantile
OR <- p_case_quantile/p_cont_quantile
OR <- OR/OR[1]
# Return output
out <- cbind(ul_qv_PRS[,3:4],p_cont_quantile, p_case_quantile, OR)
row.names(out) <- 1:n_quantile
colnames(out) <- c("q_min", "q_max","p_control", "p_case", "OR")
data.frame(out)
}1.1.2 Continuous outcomes
To convert a polygenic Z-score into an absolute estimate for a trait, we must know the predicitve utility of the polygenic score (R2), and the mean and SD of the outcome in the general population. Then it is possible to estimate the mean and SD of the trait within each polygenic score quantile using a truncated norm model (currently not theory based).
Show code
# Thank you for Alex Gillet for her work developing this code.
mean_sd_quant.f <- function(PRS_R2=0.641, Outcome_mean=1, Outcome_sd=1, n_quantile=20){
### PRS quantiles with a continuous phenotype (Y)
library(tmvtnorm)
###
E_PRS = 0
SD_PRS = sqrt(1)
E_phenotype = Outcome_mean
SD_phenotype = Outcome_sd
by_quant<-1/(n_quantile)
PRS_quantile_bounds <- qnorm(p=seq(0, 1, by=by_quant), mean= E_PRS, sd= SD_PRS)
lower_PRS_vec <- PRS_quantile_bounds[1:n_quantile]
upper_PRS_vec <- PRS_quantile_bounds[2:(n_quantile+1)]
mean_vec <- c(E_phenotype, E_PRS)
sigma_mat <- matrix(sqrt(PRS_R2)*SD_phenotype*SD_PRS, nrow=2, ncol=2)
sigma_mat[1,1] <- SD_phenotype^2
sigma_mat[2,2] <- SD_PRS^2
### mean of phenotype within the truncated PRS distribution
out_mean_Y <- rep(0, n_quantile)
### SD of phenotype within the truncated PRS distribution
out_SD_Y <- rep(0, n_quantile)
### cov of Y and PRS given truncation on PRS
out_cov_Y_PRS <- rep(0, n_quantile)
### SD of PRS given truncation on PRS
out_SD_PRS <- rep(0, n_quantile)
### mean PRS given truncation on PRS
out_mean_PRS <- rep(0, n_quantile)
for(i in 1:n_quantile){
distribution_i <- mtmvnorm(mean = mean_vec,
sigma = sigma_mat,
lower = c(-Inf, lower_PRS_vec[i]),
upper = c(Inf, upper_PRS_vec[i]),
doComputeVariance=TRUE,
pmvnorm.algorithm=GenzBretz())
out_mean_Y[i] <- distribution_i$tmean[1]
out_mean_PRS[i] <- distribution_i$tmean[2]
out_SD_Y[i] <- sqrt(distribution_i$tvar[1,1])
out_SD_PRS[i] <- sqrt(distribution_i$tvar[2,2])
out_cov_Y_PRS[i] <- distribution_i$tvar[1,2]
}
out<-data.frame(q=1:n_quantile,
q_min=lower_PRS_vec,
q_max=upper_PRS_vec,
x_mean=out_mean_Y,
x_sd=out_SD_Y)
return(out)
out_mean_Y
out_SD_Y
out_mean_PRS
out_SD_PRS
out_cov_Y_PRS
}
library(tmvtnorm)
# Create alternative of script that doesn't require simulation
mean_sd_quant.f <- function(PRS_R2=0.641, Outcome_mean=1, Outcome_sd=1, n_quantile=20){
### PRS quantiles with a continuous phenotype (Y)
library(tmvtnorm)
###
E_PRS = 0
SD_PRS = sqrt(1)
E_phenotype = Outcome_mean
SD_phenotype = Outcome_sd
n_quantile=20
by_quant<-1/(n_quantile)
PRS_quantile_bounds <- qnorm(p=seq(0, 1, by=by_quant), mean= E_PRS, sd= SD_PRS)
lower_PRS_vec <- PRS_quantile_bounds[1:n_quantile]
upper_PRS_vec <- PRS_quantile_bounds[2:(n_quantile+1)]
mean_vec <- c(E_phenotype, E_PRS)
sigma_mat <- matrix(sqrt(PRS_R2)*SD_phenotype*SD_PRS, nrow=2, ncol=2)
sigma_mat[1,1] <- SD_phenotype^2
sigma_mat[2,2] <- SD_PRS^2
### mean of phenotype within the truncated PRS distribution
out_mean_Y <- rep(0, 20)
### SD of phenotype within the truncated PRS distribution
out_SD_Y <- rep(0, 20)
### cov of Y and PRS given truncation on PRS
out_cov_Y_PRS <- rep(0, 20)
### SD of PRS given truncation on PRS
out_SD_PRS <- rep(0, 20)
### mean PRS given truncation on PRS
out_mean_PRS <- rep(0, 20)
for(i in 1:n_quantile){
distribution_i <- mtmvnorm(mean = mean_vec,
sigma = sigma_mat,
lower = c(-Inf, lower_PRS_vec[i]),
upper = c(Inf, upper_PRS_vec[i]),
doComputeVariance=TRUE,
pmvnorm.algorithm=GenzBretz())
out_mean_Y[i] <- distribution_i$tmean[1]
out_mean_PRS[i] <- distribution_i$tmean[2]
out_SD_Y[i] <- sqrt(distribution_i$tvar[1,1])
out_SD_PRS[i] <- sqrt(distribution_i$tvar[2,2])
out_cov_Y_PRS[i] <- distribution_i$tvar[1,2]
}
out<-data.frame(q=1:n_quantile,
q_min=lower_PRS_vec,
q_max=upper_PRS_vec,
x_mean=out_mean_Y,
x_sd=out_SD_Y)
return(out)
out_mean_Y
out_SD_Y
out_mean_PRS
out_SD_PRS
out_cov_Y_PRS
}
pdf('~/comp_stand.pdf')
for(i in seq(0.05, 0.95, 0.05)){
sim_res<-PRS_abs_quant2(PRS_R2 = i, Outcome_mean = 0, Outcome_sd = 1, n_quantile = 20)
nosim_res<-mean_sd_quant.f(PRS_R2 = i, Outcome_mean = 0, Outcome_sd = 1, n_quantile = 20)
plot(sim_res$x_mean,nosim_res$x_mean, main=paste0('R2 = ',i))
abline(coef = c(0,1))
}
dev.off()
pdf('~/comp_centre.pdf')
for(i in seq(0.05, 0.95, 0.05)){
sim_res<-PRS_abs_quant2(PRS_R2 = i, Outcome_mean = 0, Outcome_sd = 2, n_quantile = 20)
nosim_res<-mean_sd_quant.f(PRS_R2 = i, Outcome_mean = 0, Outcome_sd = 2, n_quantile = 20)
plot(sim_res$x_mean,nosim_res$x_mean, main=paste0('R2 = ',i))
abline(coef = c(0,1))
plot(sim_res$x_sd,nosim_res$x_sd, main=paste0('R2 = ',i))
abline(coef = c(0,1))
}
dev.off()
pdf('~/comp_scaled.pdf')
for(i in seq(0.05, 0.95, 0.05)){
sim_res<-PRS_abs_quant2(PRS_R2 = i, Outcome_mean = 100, Outcome_sd = 1, n_quantile = 20)
nosim_res<-mean_sd_quant.f(PRS_R2 = i, Outcome_mean = 100, Outcome_sd = 1, n_quantile = 20)
plot(sim_res$x_mean,nosim_res$x_mean, main=paste0('R2 = ',i))
abline(coef = c(0,1))
}
dev.off()1.2 Validate conversion
1.2.1 Calculate polygenic scores
pT + clump: Sparse
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer Depression)
gwas=$(echo DEPR07 COLL01 BODY04 HEIG03 DIAB05 COAD01 INFB01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir -p ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 5G
#SBATCH -J pt_clump
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_interpretation/1KG_ref/pt_clump/sbatch.sh
DBSLMM
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer Depression)
gwas=$(echo DEPR07 COLL01 BODY04 HEIG03 DIAB05 COAD01 INFB01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir -p ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.DBSLMM_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 5G
#SBATCH -J DBSLMM
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_DBSLMM/Scaled_polygenic_scorer_DBSLMM.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/DBSLMM/${gwas}/1KGPhase3.w_hm3.${gwas}.dbslmm.GW.txt \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/DBSLMM/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_interpretation/1KG_ref/DBSLMM/sbatch.sh
1.2.2 Evaluate polygenic scores
Compare all methods
# Create a file listing the predictors files
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
pheno<-c('Depression','Intelligence','BMI','Height','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas<-c('DEPR07','COLL01','BODY04','HEIG03','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
for(i in 1:length(pheno)){
pred_file<-NULL
# pT+clump (sparse)
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/pt_clump/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.profiles'),
group='pT+clump'))
# DBSLMM
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/DBSLMM/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.DBSLMM_profiles'),
group='DBSLMM'))
# Write out list of predictors with groups
dir.create(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',pheno[i]))
write.table(pred_file, paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',pheno[i],'/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'), row.names=F, col.names=T, quote=F)
}. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Run Model_builder_V2.R
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR07 COLL01 BODY04 HEIG03 DIAB05 COAD01 INFB01 SCLE03 RHEU02 BRCA01 PRCA01)
prev=$(echo 0.15 NA NA NA 0.05 0.03 0.013 0.00164 0.005 0.125 0.125)
# 1KG reference
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/${pheno_i}/UKBB.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp \
--n_core 1 \
--compare_predictors F \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/${pheno_i}/UKBB.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp.predictor_groups
done
1.2.3 Binary outcomes
Calculate reference-standardised polygenic scores within UK Biobank for a range of dichotomous phenotypes. Estimate the AUC/R2 of the polygenic scores in UKB. Compare measured and estimated absolute risk per PRS quantile. Use the PRScs fully baysian (pseudovalidation) polygenic scores, as this method provides a single score with good relative performance compared to other approaches.
Reference-standardised polygenic scores have already been calculated in UKB for the PRS methods comparison study, and the AUC has already been estimated. Read in polygenic scores and observed phenotype for UKB, measure proportion of cases per PRS quantile, and then estimate proportion of cases per PRS quantile.
1.2.3.1 DBSLMM
Show code
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
ccprobs.f <- function(PRS_auc=0.641, prev=0.7463, n_quantile=20){
# Convert AUC into cohen's d
d <- sqrt(2)*qnorm(PRS_auc)
# Set mean difference between cases and control polygenic scores
mu_case <- d
mu_control <- 0
# Estimate mean and variance of polygenic scores across case and control
varPRS <- prev*(1+(d^2) - (d*prev)^2) + (1-prev)*(1 - (d*prev)^2)
E_PRS <- d*prev
# Estimate polygenic score quantiles
by_quant<-1/n_quantile
p_quant <- seq(by_quant, 1-by_quant, by=by_quant)
quant_vals_PRS <- rep(0, length(p_quant))
quant_f_solve <- function(x, prev, d, pq){prev*pnorm(x-d) + (1-prev)*pnorm(x) - pq}
for(i in 1:length(p_quant)){
quant_vals_PRS[i] <- unlist(uniroot(quant_f_solve, prev=prev, d=d, pq= p_quant[i], interval=c(-2.5, 2.5), extendInt = "yes", tol=6e-12)$root)
}
# Create a table for output
ul_qv_PRS <- matrix(0, ncol=2, nrow=n_quantile)
ul_qv_PRS[1,1] <- -Inf
ul_qv_PRS[2:n_quantile,1] <- quant_vals_PRS
ul_qv_PRS[1:(n_quantile-1),2] <- quant_vals_PRS
ul_qv_PRS[n_quantile,2] <- Inf
ul_qv_PRS<-cbind(ul_qv_PRS, (ul_qv_PRS[,1:2]-E_PRS)/sqrt(varPRS))
# Estimate case control proportion for each quantile
prob_quantile_case <- pnorm(ul_qv_PRS[,2], mean = mu_case) - pnorm(ul_qv_PRS[,1], mean = mu_case)
prob_quantile_control <- pnorm(ul_qv_PRS[,2], mean = mu_control) - pnorm(ul_qv_PRS[,1], mean = mu_control)
p_case_quantile <- (prob_quantile_case*prev)/by_quant
p_cont_quantile <- (prob_quantile_control*(1-prev))/by_quant
# Estimate OR comparing each quantile to bottom quantile
OR <- p_case_quantile/p_cont_quantile
OR <- OR/OR[1]
# Return output
out <- cbind(ul_qv_PRS[,3:4],p_cont_quantile, p_case_quantile, OR)
row.names(out) <- 1:n_quantile
colnames(out) <- c("q_min", "q_max","p_control", "p_case", "OR")
data.frame(out)
}
# Run analysis for each phenotype
res_all<-NULL
cor_res<-NULL
plots_all<-list()
prs_dist_all<-list()
mean_sd<-NULL
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/DBSLMM/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.DBSLMM_profiles'))
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_DBSLMM')), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
mean_sd<-rbind(mean_sd,data.frame(Phenotype=files$pheno[i],
Mean_all=mean(pheno_prs$prs),
SD_all=sd(pheno_prs$prs),
Mean_con=mean(pheno_prs$prs[pheno_prs$pheno == 0]),
SD_con=sd(pheno_prs$prs[pheno_prs$pheno == 0]),
Mean_cas=mean(pheno_prs$prs[pheno_prs$pheno == 1]),
SD_cas=sd(pheno_prs$prs[pheno_prs$pheno == 1]),
f_test_pval=var.test(prs ~ pheno, pheno_prs, alternative = "two.sided")$p.value))
# Plot DBSLMM PRS distribution
library(ggplot2)
library(cowplot)
prs_dist_all[[i]]<-ggplot(pheno_prs, aes(x=prs)) +
geom_histogram() +
labs(y="Count", x='Polygenic Score', title=files$pheno[i]) +
theme_cowplot(12)
# Read in AUC for PRS
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',files$pheno[i],'/UKBB.w_hm3.',files$gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
prs_auc<-assoc[grepl('DBSLMM', assoc$Predictor),]$AUC
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs$obs_quant<-as.numeric(cut( pheno_prs$prs, obs_quant, include.lowest = T))
# Calculate proportion of each quantile that are cases
obs_cc<-NULL
for(k in 1:n_quant){
obs_cc<-rbind(obs_cc, data.frame(Phenotype=files$pheno[i],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
p_control=1-mean(pheno_prs$pheno[pheno_prs$obs_quant == k]),
p_case=mean(pheno_prs$pheno[pheno_prs$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_cc<-ccprobs.f(PRS_auc = prs_auc, prev=mean(pheno_prs$pheno), n_quantile = n_quant)
est_cc$OR<-NULL
est_cc<-data.frame(Phenotype=files$pheno[i],Type="\nEstimated\n(Observed AUC)",Quantile=1:n_quant, est_cc)
est_quant<-sort(unique(c(est_cc$q_min, est_cc$q_max)))
pheno_prs$est_quant<-as.numeric(cut( pheno_prs$prs, est_quant, include.lowest = T))
tmp<-cor.test(obs_cc$p_case, est_cc$p_case)
tmp2<-abs(est_cc$p_case-obs_cc$p_case)/obs_cc$p_case
# Estimate correlation between observed and expected
cor_res<-rbind(cor_res,data.frame(Phenotype=files$pheno[i],
Cor=tmp$estimate,
Low95CI=tmp$conf.int[1],
High95CI=tmp$conf.int[2],
Mean_perc_diff=mean(tmp2),
N=length(pheno_prs$pheno),
Ncas=sum(pheno_prs$pheno == 1),
Ncon=sum(pheno_prs$pheno == 0)))
quant_comp<-rbind(obs_cc, est_cc)
res_all<-rbind(res_all, quant_comp)
library(ggplot2)
library(cowplot)
plots_all[[i]]<-ggplot(quant_comp, aes(x=Quantile, y=p_case, colour=Type)) +
geom_point(alpha=0.8) +
geom_line(alpha=0.8) +
labs(y="p(case)", title=files$pheno[i], colour='Method') +
theme_cowplot(12)
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PRS_dist_binary.png'), units='px', res=300, width=2000, height=2800)
plot_grid(plotlist=prs_dist_all, ncol = 2)
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PropCC_Comp.png'), units='px', res=300, width=2000, height=2800)
plot_grid(plotlist=plots_all, ncol = 2)
dev.off()
write.csv(cor_res, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PropCC_Comp.csv', row.names=F, quote=F)
write.csv(mean_sd, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PRS_Mean_SD.csv', row.names=F, quote=F)Show results
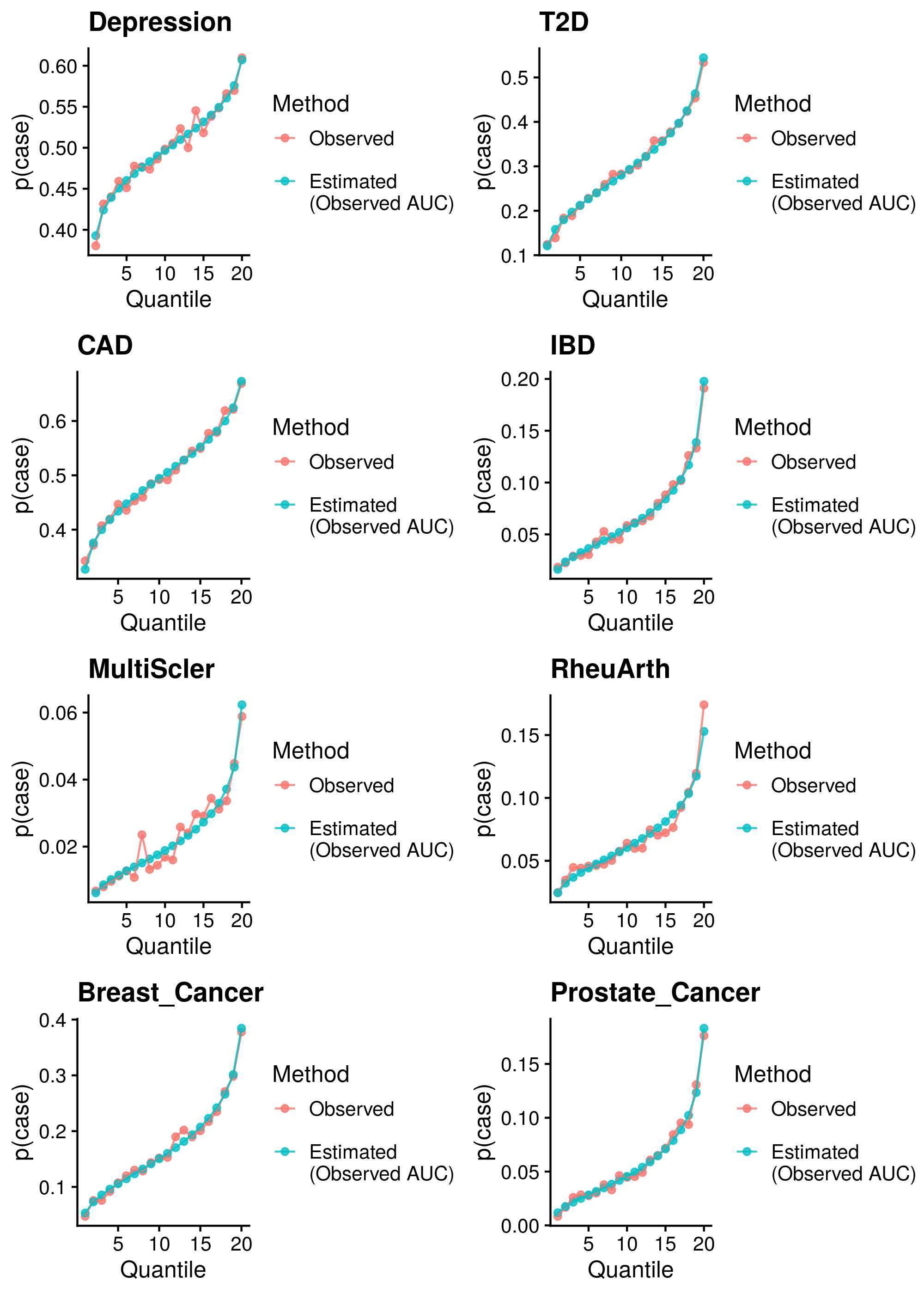
| Phenotype | Correlation (95%CI) | Mean Abs. Diff. | N | Ncas | Ncon |
|---|---|---|---|---|---|
| Depression | 0.985 (0.961-0.994) | 1.5% | 49999 | 24999 | 25000 |
| T2D | 0.997 (0.992-0.999) | 2.4% | 49999 | 14888 | 35111 |
| CAD | 0.994 (0.985-0.998) | 1.5% | 49999 | 25000 | 24999 |
| IBD | 0.994 (0.985-0.998) | 6.8% | 49999 | 3461 | 46538 |
| MultiScler | 0.969 (0.922-0.988) | 12.6% | 49999 | 1137 | 48862 |
| RheuArth | 0.981 (0.952-0.993) | 6.8% | 49999 | 3408 | 46591 |
| Breast_Cancer | 0.995 (0.987-0.998) | 4.6% | 49999 | 8512 | 41487 |
| Prostate_Cancer | 0.994 (0.984-0.998) | 8.7% | 50000 | 2927 | 47073 |
Median Cor. = 0.994053; Mean Cor. = 0.9886512; Min. Cor. = 0.9691668; Max. Cor. = 0.9968126 Mean Abs. Diff = 0.05621957; Min. Mean Abs. Diff. = 0.01504855; Max. Mean Abs. Diff. = 0.1262171
1.2.3.2 pT+clump
Show code
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
ccprobs.f <- function(PRS_auc=0.641, prev=0.7463, n_quantile=20){
# Convert AUC into cohen's d
d <- sqrt(2)*qnorm(PRS_auc)
# Set mean difference between cases and control polygenic scores
mu_case <- d
mu_control <- 0
# Estimate mean and variance of polygenic scores across case and control
varPRS <- prev*(1+(d^2) - (d*prev)^2) + (1-prev)*(1 - (d*prev)^2)
E_PRS <- d*prev
# Estimate polygenic score quantiles
by_quant<-1/n_quantile
p_quant <- seq(by_quant, 1-by_quant, by=by_quant)
quant_vals_PRS <- rep(0, length(p_quant))
quant_f_solve <- function(x, prev, d, pq){prev*pnorm(x-d) + (1-prev)*pnorm(x) - pq}
for(i in 1:length(p_quant)){
quant_vals_PRS[i] <- unlist(uniroot(quant_f_solve, prev=prev, d=d, pq= p_quant[i], interval=c(-2.5, 2.5), extendInt = "yes", tol=6e-12)$root)
}
# Create a table for output
ul_qv_PRS <- matrix(0, ncol=2, nrow=n_quantile)
ul_qv_PRS[1,1] <- -Inf
ul_qv_PRS[2:n_quantile,1] <- quant_vals_PRS
ul_qv_PRS[1:(n_quantile-1),2] <- quant_vals_PRS
ul_qv_PRS[n_quantile,2] <- Inf
ul_qv_PRS<-cbind(ul_qv_PRS, (ul_qv_PRS[,1:2]-E_PRS)/sqrt(varPRS))
# Estimate case control proportion for each quantile
prob_quantile_case <- pnorm(ul_qv_PRS[,2], mean = mu_case) - pnorm(ul_qv_PRS[,1], mean = mu_case)
prob_quantile_control <- pnorm(ul_qv_PRS[,2], mean = mu_control) - pnorm(ul_qv_PRS[,1], mean = mu_control)
p_case_quantile <- (prob_quantile_case*prev)/by_quant
p_cont_quantile <- (prob_quantile_control*(1-prev))/by_quant
# Estimate OR comparing each quantile to bottom quantile
OR <- p_case_quantile/p_cont_quantile
OR <- OR/OR[1]
# Return output
out <- cbind(ul_qv_PRS[,3:4],p_cont_quantile, p_case_quantile, OR)
row.names(out) <- 1:n_quantile
colnames(out) <- c("q_min", "q_max","p_control", "p_case", "OR")
data.frame(out)
}
# Run analysis for each phenotype
res_all<-NULL
cor_res<-NULL
plots_all<-list()
prs_dist_all<-list()
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/pt_clump/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.profiles'))
# Extract PRS with the most stringent p-value threshold
score_nsnp<-fread(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/pt_clump/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.NSNP_per_pT'))
score_nsnp<-score_nsnp[score_nsnp$NSNP >= 5,]
nsnp<-score_nsnp$NSNP[score_nsnp$pT1 == min(score_nsnp$pT1)]
pT<-min(score_nsnp$pT1)
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_',pT)), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
# Plot DBSLMM PRS distribution
library(ggplot2)
library(cowplot)
prs_dist_all[[i]]<-ggplot(pheno_prs, aes(x=prs)) +
geom_histogram() +
labs(y="Count", x='Polygenic Score', title=paste0(files$pheno[i],': ',nsnp,' SNPs')) +
theme_cowplot(12)
# Read in AUC for PRS
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',files$pheno[i],'/UKBB.w_hm3.',files$gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
prs_auc<-assoc[grepl(paste0(gwas[i],'_',gsub('-','.',pT)), assoc$Predictor),]$AUC
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs$obs_quant<-as.numeric(cut( pheno_prs$prs, obs_quant, include.lowest = T))
# Calculate proportion of each quantile that are cases
obs_cc<-NULL
for(k in 1:n_quant){
obs_cc<-rbind(obs_cc, data.frame(Phenotype=files$pheno[i],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
p_control=1-mean(pheno_prs$pheno[pheno_prs$obs_quant == k]),
p_case=mean(pheno_prs$pheno[pheno_prs$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_cc<-ccprobs.f(PRS_auc = prs_auc, prev=mean(pheno_prs$pheno), n_quantile = n_quant)
est_cc$OR<-NULL
est_cc<-data.frame(Phenotype=files$pheno[i],Type="\nEstimated\n(Observed AUC)",Quantile=1:n_quant, est_cc)
est_quant<-sort(unique(c(est_cc$q_min, est_cc$q_max)))
pheno_prs$est_quant<-as.numeric(cut( pheno_prs$prs, est_quant, include.lowest = T))
tmp<-cor.test(obs_cc$p_case, est_cc$p_case)
tmp2<-abs(est_cc$p_case-obs_cc$p_case)/obs_cc$p_case
# Estimate correlation between observed and expected
cor_res<-rbind(cor_res,data.frame(Phenotype=files$pheno[i],
Cor=tmp$estimate,
Low95CI=tmp$conf.int[1],
High95CI=tmp$conf.int[2],
Mean_perc_diff=mean(tmp2),
N=length(pheno_prs$pheno),
Ncas=sum(pheno_prs$pheno == 1),
Ncon=sum(pheno_prs$pheno == 0)))
quant_comp<-rbind(obs_cc, est_cc)
res_all<-rbind(res_all, quant_comp)
library(ggplot2)
library(cowplot)
plots_all[[i]]<-ggplot(quant_comp, aes(x=Quantile, y=p_case, colour=Type)) +
geom_point(alpha=0.8) +
geom_line(alpha=0.8) +
labs(y="p(case)", title=paste0(files$pheno[i],': \n',nsnp,' SNPs; AUC=',round(prs_auc,3)), colour='Method') +
theme_cowplot(12)
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/pt_clump_PRS_dist_binary.png'), units='px', res=300, width=2000, height=2800)
plot_grid(plotlist=prs_dist_all, ncol = 2)
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/pt_clump_PropCC_Comp.png'), units='px', res=300, width=2000, height=2800)
plot_grid(plotlist=plots_all, ncol = 2)
dev.off()
write.csv(cor_res, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/pt_clump_PropCC_Comp.csv', row.names=F, quote=F)Show results
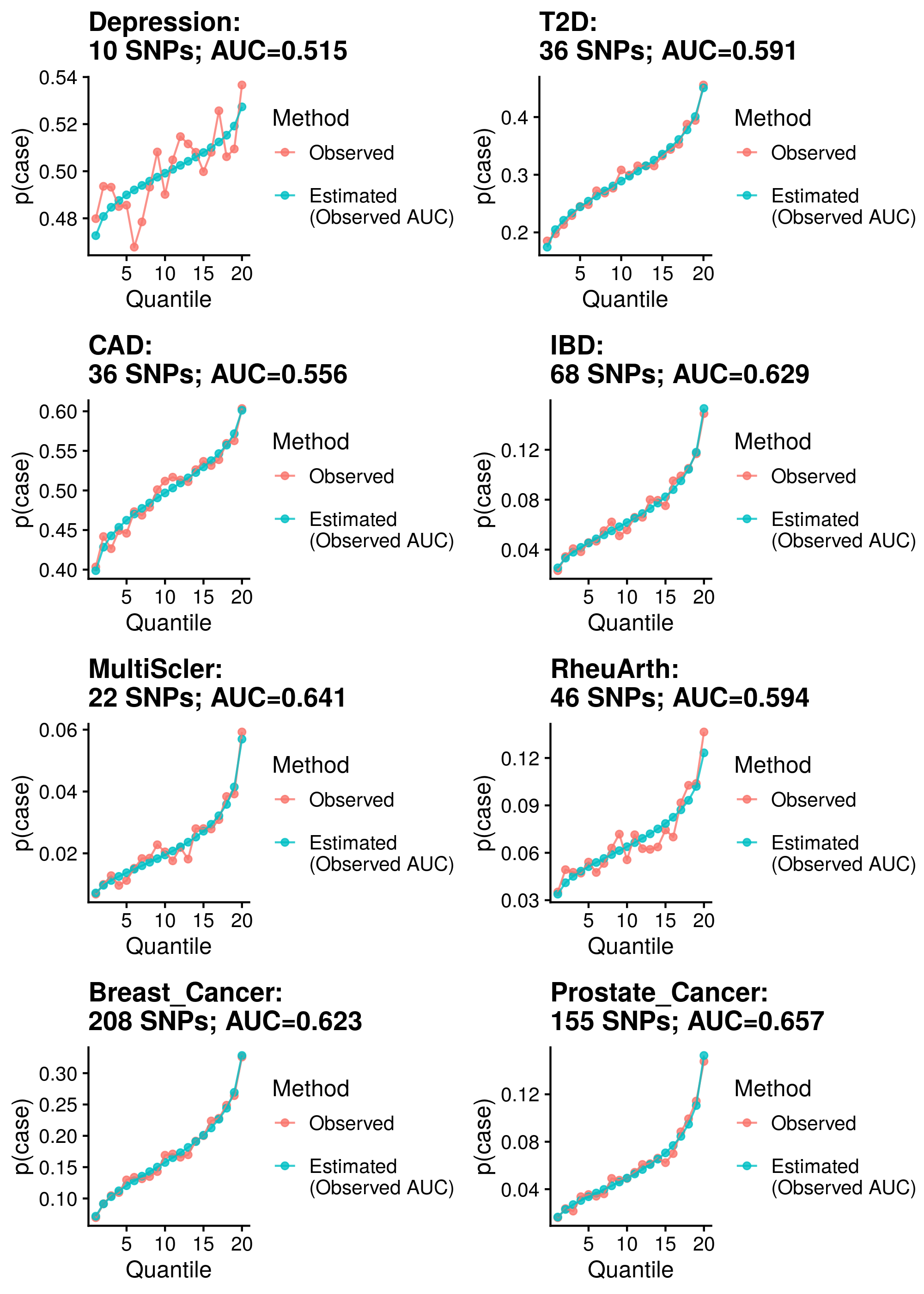
| Phenotype | Correlation (95%CI) | Mean Abs. Diff. | N | Ncas | Ncon |
|---|---|---|---|---|---|
| Depression | 0.779 (0.513-0.908) | 1.8% | 49999 | 24999 | 25000 |
| T2D | 0.993 (0.983-0.997) | 2.3% | 49999 | 14888 | 35111 |
| CAD | 0.983 (0.956-0.993) | 1.6% | 49999 | 25000 | 24999 |
| IBD | 0.99 (0.974-0.996) | 6% | 49999 | 3461 | 46538 |
| MultiScler | 0.981 (0.95-0.992) | 10.4% | 49999 | 1137 | 48862 |
| RheuArth | 0.951 (0.878-0.981) | 9.5% | 49999 | 3408 | 46591 |
| Breast_Cancer | 0.995 (0.987-0.998) | 3.3% | 49999 | 8512 | 41487 |
| Prostate_Cancer | 0.992 (0.98-0.997) | 6.6% | 50000 | 2927 | 47073 |
Median Cor. = 0.9863451; Mean Cor. = 0.9579612; Min. Cor. = 0.7786503; Max. Cor. = 0.9948055
1.2.4 Continuous outcomes
Calculate reference-standardised polygenic scores within UK Biobank for a range of continuous phenotypes. Estimate the R2 of the polygenic scores in UKB. Compare measured and estimated absolute meana and sd per PRS quantile. Use the DBSLMM fully baysian (pseudovalidation) polygenic scores, as this method provides a single score with good relative performance compared to other approaches.
Reference-standardised polygenic scores have already been calculated in UKB for the PRS methods comparison study, and the R2 has already been estimated. Read in polygenic scores and observed phenotype for UKB, measure phenotype mean and sd per PRS quantile, and then estimate measure phenotype mean and sd per PRS quantile.
1.2.4.1 DBSLMM
Show code
library(data.table)
library(e1071)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
gwas<-c('COLL01','HEIG03','BODY04')
pheno<-c('Intelligence','Height','BMI')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
mean_sd_quant.f <- function(PRS_R2=0.641, Outcome_mean=1, Outcome_sd=1, n_quantile=20){
### PRS quantiles with a continuous phenotype (Y)
library(tmvtnorm)
###
E_PRS = 0
SD_PRS = sqrt(1)
E_phenotype = Outcome_mean
SD_phenotype = Outcome_sd
by_quant<-1/(n_quantile)
PRS_quantile_bounds <- qnorm(p=seq(0, 1, by=by_quant), mean= E_PRS, sd= SD_PRS)
lower_PRS_vec <- PRS_quantile_bounds[1:n_quantile]
upper_PRS_vec <- PRS_quantile_bounds[2:(n_quantile+1)]
mean_vec <- c(E_phenotype, E_PRS)
sigma_mat <- matrix(sqrt(PRS_R2)*SD_phenotype*SD_PRS, nrow=2, ncol=2)
sigma_mat[1,1] <- SD_phenotype^2
sigma_mat[2,2] <- SD_PRS^2
### mean of phenotype within the truncated PRS distribution
out_mean_Y <- rep(0, n_quantile)
### SD of phenotype within the truncated PRS distribution
out_SD_Y <- rep(0, n_quantile)
### cov of Y and PRS given truncation on PRS
out_cov_Y_PRS <- rep(0, n_quantile)
### SD of PRS given truncation on PRS
out_SD_PRS <- rep(0, n_quantile)
### mean PRS given truncation on PRS
out_mean_PRS <- rep(0, n_quantile)
for(i in 1:n_quantile){
distribution_i <- mtmvnorm(mean = mean_vec,
sigma = sigma_mat,
lower = c(-Inf, lower_PRS_vec[i]),
upper = c(Inf, upper_PRS_vec[i]),
doComputeVariance=TRUE,
pmvnorm.algorithm=GenzBretz())
out_mean_Y[i] <- distribution_i$tmean[1]
out_mean_PRS[i] <- distribution_i$tmean[2]
out_SD_Y[i] <- sqrt(distribution_i$tvar[1,1])
out_SD_PRS[i] <- sqrt(distribution_i$tvar[2,2])
out_cov_Y_PRS[i] <- distribution_i$tvar[1,2]
}
out<-data.frame(q=1:n_quantile,
q_min=lower_PRS_vec,
q_max=upper_PRS_vec,
x_mean=out_mean_Y,
x_sd=out_SD_Y)
return(out)
out_mean_Y
out_SD_Y
out_mean_PRS
out_SD_PRS
out_cov_Y_PRS
}
# Run analysis for each phenotype
res_all<-NULL
plots_all<-list()
cor_res<-NULL
prs_dist_all<-list()
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/DBSLMM/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.DBSLMM_profiles'))
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_DBSLMM')), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
# Plot DBSLMM PRS distribution
library(ggplot2)
library(cowplot)
prs_dist_all[[i]]<-ggplot(pheno_prs, aes(x=prs)) +
geom_histogram() +
labs(y="Count", x='Polygenic Score', title=files$pheno[i]) +
theme_cowplot(12)
# Read in AUC for PRS
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',files$pheno[i],'/UKBB.w_hm3.',files$gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
prs_r2<-assoc[grepl('DBSLMM', assoc$Predictor),]$Obs_R2
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs$obs_quant<-as.numeric(cut( pheno_prs$prs, obs_quant, include.lowest = T))
# Calculate mean and SD of each quantile that are cases
obs_dist<-NULL
for(k in 1:n_quant){
obs_dist<-rbind(obs_dist, data.frame(Phenotype=files$pheno[i],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
x_mean=mean(pheno_prs$pheno[pheno_prs$obs_quant == k]),
x_sd=sd(pheno_prs$pheno[pheno_prs$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_dist<-mean_sd_quant.f(PRS_R2 = prs_r2, Outcome_mean=mean(pheno_prs$pheno), Outcome_sd=sd(pheno_prs$pheno), n_quantile = n_quant)
est_dist$q<-NULL
est_dist<-data.frame(Phenotype=files$pheno[i],Type="\nEstimated\n(Observed R2)",Quantile=1:n_quant, est_dist)
est_quant<-sort(unique(c(est_dist$q_min, est_dist$q_max)))
pheno_prs$est_quant<-as.numeric(cut( pheno_prs$prs, est_quant, include.lowest = T))
quant_comp<-rbind(obs_dist, est_dist)
tmp<-cor.test(obs_dist$x_mean, est_dist$x_mean)
tmp2<-abs(est_dist$x_mean-obs_dist$x_mean)/obs_dist$x_mean
cor_res<-rbind(cor_res,data.frame(Phenotype=files$pheno[i],
Cor_mean=tmp$estimate,
Cor_mean_Low95CI=tmp$conf.int[1],
Cor_mean_High95CI=tmp$conf.int[2],
Mean_perc_diff_mean=mean(abs(est_dist$x_mean-obs_dist$x_mean)/obs_dist$x_mean),
Mean_perc_diff_sd=mean(abs(est_dist$x_sd-obs_dist$x_sd)/obs_dist$x_sd),
N=length(pheno_prs$pheno),
Skewness=skewness(pheno_prs$pheno)))
res_all<-rbind(res_all, quant_comp)
library(ggplot2)
library(cowplot)
plots_all[[i]]<-ggplot(quant_comp, aes(x=Quantile, y=x_mean, colour=Type)) +
geom_point(stat="identity", position=position_dodge(.5), alpha=0.8, shape=18, size=3) +
geom_errorbar(aes(ymin=x_mean-x_sd, ymax=x_mean+x_sd), width=.2, position=position_dodge(.5), alpha=0.8) +
labs(y="Mean (SD)", title=files$pheno[i], colour='Method') +
theme_cowplot(12)
# geom_vline(xintercept = seq(1.5,19.5,1), linetype="dotted", color = "black")
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PRS_dist_cont.png'), units='px', res=300, width=1750, height=2000)
plot_grid(plotlist=prs_dist_all, ncol = 1)
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/Mean_SD_Comp.png'), units='px', res=300, width=1750, height=2000)
plot_grid(plotlist=plots_all, ncol = 1)
dev.off()
write.csv(cor_res, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/Mean_SD_Comp.csv', row.names=F, quote=F)Show results
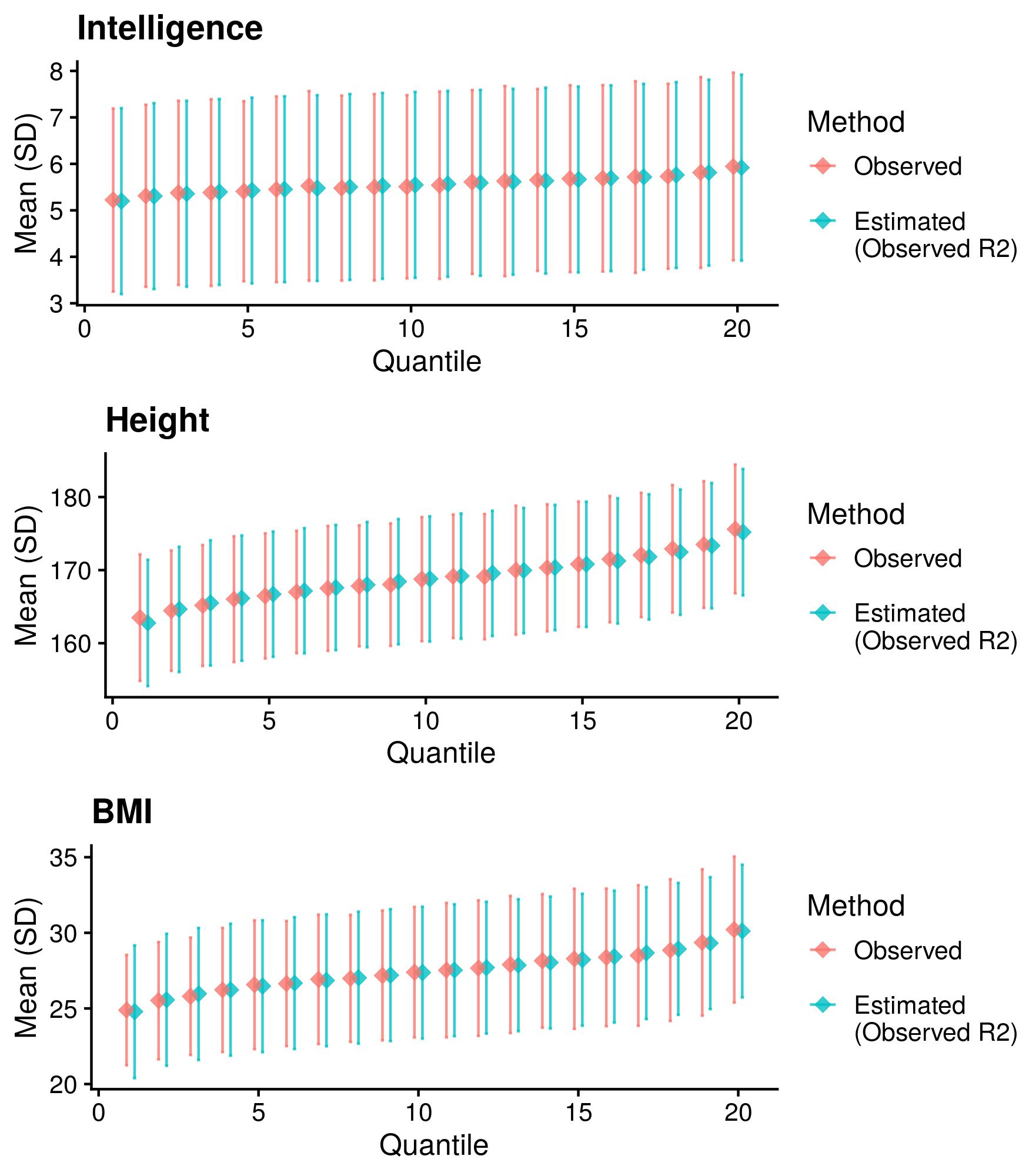
| Phenotype | Correlation (95%CI) | Mean Abs. Diff. of Mean | Mean Abs. Diff. of SD | N | Skewness |
|---|---|---|---|---|---|
| Intelligence | 0.992 (0.979-0.997) | 0.3% | 1.3% | 50000 | 0.144 |
| Height | 0.996 (0.989-0.998) | 0.1% | 1.5% | 49999 | 0.117 |
| BMI | 0.998 (0.995-0.999) | 0.2% | 6% | 49999 | 0.592 |
Median Cor. of means = 0.9958381; Mean Cor. of means = 0.9952452; Min. Cor. of means = 0.9917264; Max. Cor. of means = 0.9981713
## Warning in mean.default(res$PercDiff_sd_mean): argument is not numeric or
## logical: returning NA## Warning in min(res$PercDiff_sd_mean): no non-missing arguments to min; returning
## Inf## Warning in max(res$PercDiff_sd_mean): no non-missing arguments to max; returning
## -InfMedian mean %diff of SD = ; Mean mean %diff of SD = NA; Min. mean %diff of SD = Inf; Max. mean %diff of SD = -Inf
1.2.4.2 pt+clump
Show code
library(data.table)
library(e1071)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
gwas<-c('COLL01','HEIG03','BODY04')
pheno<-c('Intelligence','Height','BMI')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
mean_sd_quant.f <- function(PRS_R2=0.641, Outcome_mean=1, Outcome_sd=1, n_quantile=20){
### PRS quantiles with a continuous phenotype (Y)
library(tmvtnorm)
###
E_PRS = 0
SD_PRS = sqrt(1)
E_phenotype = Outcome_mean
SD_phenotype = Outcome_sd
by_quant<-1/(n_quantile)
PRS_quantile_bounds <- qnorm(p=seq(0, 1, by=by_quant), mean= E_PRS, sd= SD_PRS)
lower_PRS_vec <- PRS_quantile_bounds[1:n_quantile]
upper_PRS_vec <- PRS_quantile_bounds[2:(n_quantile+1)]
mean_vec <- c(E_phenotype, E_PRS)
sigma_mat <- matrix(sqrt(PRS_R2)*SD_phenotype*SD_PRS, nrow=2, ncol=2)
sigma_mat[1,1] <- SD_phenotype^2
sigma_mat[2,2] <- SD_PRS^2
### mean of phenotype within the truncated PRS distribution
out_mean_Y <- rep(0, n_quantile)
### SD of phenotype within the truncated PRS distribution
out_SD_Y <- rep(0, n_quantile)
### cov of Y and PRS given truncation on PRS
out_cov_Y_PRS <- rep(0, n_quantile)
### SD of PRS given truncation on PRS
out_SD_PRS <- rep(0, n_quantile)
### mean PRS given truncation on PRS
out_mean_PRS <- rep(0, n_quantile)
for(i in 1:n_quantile){
distribution_i <- mtmvnorm(mean = mean_vec,
sigma = sigma_mat,
lower = c(-Inf, lower_PRS_vec[i]),
upper = c(Inf, upper_PRS_vec[i]),
doComputeVariance=TRUE,
pmvnorm.algorithm=GenzBretz())
out_mean_Y[i] <- distribution_i$tmean[1]
out_mean_PRS[i] <- distribution_i$tmean[2]
out_SD_Y[i] <- sqrt(distribution_i$tvar[1,1])
out_SD_PRS[i] <- sqrt(distribution_i$tvar[2,2])
out_cov_Y_PRS[i] <- distribution_i$tvar[1,2]
}
out<-data.frame(q=1:n_quantile,
q_min=lower_PRS_vec,
q_max=upper_PRS_vec,
x_mean=out_mean_Y,
x_sd=out_SD_Y)
return(out)
out_mean_Y
out_SD_Y
out_mean_PRS
out_SD_PRS
out_cov_Y_PRS
}
# Run analysis for each phenotype
res_all<-NULL
plots_all<-list()
cor_res<-NULL
prs_dist_all<-list()
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/pt_clump/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.profiles'))
score_nsnp<-fread(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/pt_clump/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.NSNP_per_pT'))
score_nsnp<-score_nsnp[score_nsnp$NSNP >= 5,]
nsnp<-score_nsnp$NSNP[score_nsnp$pT1 == min(score_nsnp$pT1)]
pT<-min(score_nsnp$pT1)
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_',pT)), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
# Plot PRS distribution
library(ggplot2)
library(cowplot)
prs_dist_all[[i]]<-ggplot(pheno_prs, aes(x=prs)) +
geom_histogram() +
labs(y="Count", x='Polygenic Score', title=paste0(files$pheno[i],': ',nsnp,' SNPs')) +
theme_cowplot(12)
# Read in R2 for PRS
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',files$pheno[i],'/UKBB.w_hm3.',files$gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
prs_r2<-assoc[grepl(paste0(gwas[i],'_',gsub('-','.',pT)), assoc$Predictor),]$Obs_R2
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs$obs_quant<-as.numeric(cut( pheno_prs$prs, obs_quant, include.lowest = T))
# Calculate mean and SD of each quantile
obs_dist<-NULL
for(k in 1:n_quant){
obs_dist<-rbind(obs_dist, data.frame(Phenotype=files$pheno[i],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
x_mean=mean(pheno_prs$pheno[pheno_prs$obs_quant == k]),
x_sd=sd(pheno_prs$pheno[pheno_prs$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_dist<-mean_sd_quant.f(PRS_R2 = prs_r2, Outcome_mean=mean(pheno_prs$pheno), Outcome_sd=sd(pheno_prs$pheno), n_quantile = n_quant)
est_dist$q<-NULL
est_dist<-data.frame(Phenotype=files$pheno[i],Type="\nEstimated\n(Observed R2)",Quantile=1:n_quant, est_dist)
est_quant<-sort(unique(c(est_dist$q_min, est_dist$q_max)))
pheno_prs$est_quant<-as.numeric(cut( pheno_prs$prs, est_quant, include.lowest = T))
quant_comp<-rbind(obs_dist, est_dist)
tmp<-cor.test(obs_dist$x_mean, est_dist$x_mean)
tmp2<-abs(est_dist$x_mean-obs_dist$x_mean)/obs_dist$x_mean
cor_res<-rbind(cor_res,data.frame(Phenotype=files$pheno[i],
Cor_mean=tmp$estimate,
Cor_mean_Low95CI=tmp$conf.int[1],
Cor_mean_High95CI=tmp$conf.int[2],
Mean_perc_diff_mean=mean(abs(est_dist$x_mean-obs_dist$x_mean)/obs_dist$x_mean),
Mean_perc_diff_sd=mean(abs(est_dist$x_sd-obs_dist$x_sd)/obs_dist$x_sd),
N=length(pheno_prs$pheno),
Skewness=skewness(pheno_prs$pheno)))
res_all<-rbind(res_all, quant_comp)
library(ggplot2)
library(cowplot)
plots_all[[i]]<-ggplot(quant_comp, aes(x=Quantile, y=x_mean, colour=Type)) +
geom_point(stat="identity", position=position_dodge(.5), alpha=0.8, shape=18, size=3) +
geom_errorbar(aes(ymin=x_mean-x_sd, ymax=x_mean+x_sd), width=.2, position=position_dodge(.5), alpha=0.8) +
labs(y="Mean (SD)", title=paste0(files$pheno[i],':\n',nsnp,' SNPs; R2=',round(prs_r2,3)), colour='Method') +
theme_cowplot(12)
# geom_vline(xintercept = seq(1.5,19.5,1), linetype="dotted", color = "black")
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/pt_clump_PRS_dist_cont.png'), units='px', res=300, width=1750, height=2000)
plot_grid(plotlist=prs_dist_all, ncol = 1)
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/pt_clump_Mean_SD_Comp.png'), units='px', res=300, width=1750, height=2000)
plot_grid(plotlist=plots_all, ncol = 1)
dev.off()
write.csv(cor_res, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/pt_clump_Mean_SD_Comp.csv', row.names=F, quote=F)Show results

| Phenotype | Correlation (95%CI) | Mean Abs. Diff. of Mean | Mean Abs. Diff. of SD | N | Skewness |
|---|---|---|---|---|---|
| Intelligence | 0.992 (0.979-0.997) | 0.3% | 1.3% | 50000 | 0.144 |
| Height | 0.996 (0.989-0.998) | 0.1% | 1.5% | 49999 | 0.117 |
| BMI | 0.998 (0.995-0.999) | 0.2% | 6% | 49999 | 0.592 |
Median Cor. of means = 0.9958381; Mean Cor. of means = 0.9952452; Min. Cor. of means = 0.9917264; Max. Cor. of means = 0.9981713
## Warning in mean.default(res$PercDiff_sd_mean): argument is not numeric or
## logical: returning NA## Warning in min(res$PercDiff_sd_mean): no non-missing arguments to min; returning
## Inf## Warning in max(res$PercDiff_sd_mean): no non-missing arguments to max; returning
## -InfMedian mean %diff of SD = ; Mean mean %diff of SD = NA; Min. mean %diff of SD = Inf; Max. mean %diff of SD = -Inf
1.2.4.3 Sex stratified
Show code
library(data.table)
library(e1071)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
gwas<-c('COLL01','HEIG03','BODY04')
pheno<-c('Intelligence','Height','BMI')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
mean_sd_quant.f <- function(PRS_R2=0.641, Outcome_mean=1, Outcome_sd=1, n_quantile=20){
### PRS quantiles with a continuous phenotype (Y)
library(tmvtnorm)
###
E_PRS = 0
SD_PRS = sqrt(1)
E_phenotype = Outcome_mean
SD_phenotype = Outcome_sd
by_quant<-1/(n_quantile)
PRS_quantile_bounds <- qnorm(p=seq(0, 1, by=by_quant), mean= E_PRS, sd= SD_PRS)
lower_PRS_vec <- PRS_quantile_bounds[1:n_quantile]
upper_PRS_vec <- PRS_quantile_bounds[2:(n_quantile+1)]
mean_vec <- c(E_phenotype, E_PRS)
sigma_mat <- matrix(sqrt(PRS_R2)*SD_phenotype*SD_PRS, nrow=2, ncol=2)
sigma_mat[1,1] <- SD_phenotype^2
sigma_mat[2,2] <- SD_PRS^2
### mean of phenotype within the truncated PRS distribution
out_mean_Y <- rep(0, n_quantile)
### SD of phenotype within the truncated PRS distribution
out_SD_Y <- rep(0, n_quantile)
### cov of Y and PRS given truncation on PRS
out_cov_Y_PRS <- rep(0, n_quantile)
### SD of PRS given truncation on PRS
out_SD_PRS <- rep(0, n_quantile)
### mean PRS given truncation on PRS
out_mean_PRS <- rep(0, n_quantile)
for(i in 1:n_quantile){
distribution_i <- mtmvnorm(mean = mean_vec,
sigma = sigma_mat,
lower = c(-Inf, lower_PRS_vec[i]),
upper = c(Inf, upper_PRS_vec[i]),
doComputeVariance=TRUE,
pmvnorm.algorithm=GenzBretz())
out_mean_Y[i] <- distribution_i$tmean[1]
out_mean_PRS[i] <- distribution_i$tmean[2]
out_SD_Y[i] <- sqrt(distribution_i$tvar[1,1])
out_SD_PRS[i] <- sqrt(distribution_i$tvar[2,2])
out_cov_Y_PRS[i] <- distribution_i$tvar[1,2]
}
out<-data.frame(q=1:n_quantile,
q_min=lower_PRS_vec,
q_max=upper_PRS_vec,
x_mean=out_mean_Y,
x_sd=out_SD_Y)
return(out)
out_mean_Y
out_SD_Y
out_mean_PRS
out_SD_PRS
out_cov_Y_PRS
}
# Run analysis for each phenotype
res_all<-NULL
plots_all<-list()
cor_res<-NULL
prs_dist_all<-list()
obs_dist<-NULL
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/DBSLMM/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.DBSLMM_profiles'))
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_DBSLMM')), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
# Split sample sex (0=female, 1=male)
ukb_sex<-fread('/users/k1806347/brc_scratch/Data/UKBB/Phenotype/UKBB_Sex.pheno')
sex_code<-data.frame(code=c(0,1), sex=c('Female', 'Male'))
pheno_prs<-merge(pheno_prs, ukb_sex, by=c('FID','IID'))
cor(pheno_prs[,-1:-2])^2 # The sum of R2 values for independent variables is equal to the R2 of a model combining all variables. PRS_R2/(1-sex_R2) ~ PRS_R2 within each sex. This logic could be used to account for additional uncorrelated factors.
obs_dist<-NULL
est_dist_all<-NULL
for(sex in 0:1){
pheno_prs_sex<-pheno_prs[pheno_prs$Sex == sex,]
prs_r2<-cor(pheno_prs_sex$pheno, pheno_prs_sex$prs)^2
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs_sex$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs_sex$obs_quant<-as.numeric(cut( pheno_prs_sex$prs, obs_quant, include.lowest = T))
# Calculate mean and SD of each quantile that are cases
for(k in 1:n_quant){
obs_dist<-rbind(obs_dist, data.frame(Phenotype=files$pheno[i],
Sex=sex_code$sex[sex_code$code == sex],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
x_mean=mean(pheno_prs_sex$pheno[pheno_prs_sex$obs_quant == k]),
x_sd=sd(pheno_prs_sex$pheno[pheno_prs_sex$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_dist<-mean_sd_quant.f(PRS_R2 = prs_r2, Outcome_mean=mean(pheno_prs_sex$pheno), Outcome_sd=sd(pheno_prs_sex$pheno), n_quantile = n_quant)
est_dist$q<-NULL
est_dist<-data.frame(Phenotype=files$pheno[i],
Sex=sex_code$sex[sex_code$code == sex], Type="\nEstimated\n(Observed R2)",Quantile=1:n_quant, est_dist)
est_quant<-sort(unique(c(est_dist$q_min, est_dist$q_max)))
pheno_prs_sex$est_quant<-as.numeric(cut( pheno_prs_sex$prs, est_quant, include.lowest = T))
est_dist_all<-rbind(est_dist_all, est_dist)
}
quant_comp<-rbind(obs_dist, est_dist_all)
library(ggplot2)
library(cowplot)
plots_all[[paste0(i)]]<-ggplot(quant_comp, aes(x=Quantile, y=x_mean, colour=Type)) +
geom_point(stat="identity", position=position_dodge(.5), alpha=0.8, shape=18, size=3) +
geom_errorbar(aes(ymin=x_mean-x_sd, ymax=x_mean+x_sd), width=.2, position=position_dodge(.5), alpha=0.8) +
labs(y="Mean (SD)", title=paste0(files$pheno[i]), colour='Method') +
theme_cowplot(12) +
facet_grid(. ~ Sex)
# geom_vline(xintercept = seq(1.5,19.5,1), linetype="dotted", color = "black")
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/Mean_SD_Comp_sex_stratified.png'), units='px', res=300, width=2000, height=2000)
plot_grid(plotlist=plots_all, ncol = 1)
dev.off()Show results

| Phenotype | Correlation (95%CI) | Mean Abs. Diff. of Mean | Mean Abs. Diff. of SD | N | Skewness |
|---|---|---|---|---|---|
| Intelligence | 0.992 (0.979-0.997) | 0.3% | 1.3% | 50000 | 0.144 |
| Height | 0.996 (0.989-0.998) | 0.1% | 1.5% | 49999 | 0.117 |
| BMI | 0.998 (0.995-0.999) | 0.2% | 6% | 49999 | 0.592 |
Median Cor. of means = 0.9958381; Mean Cor. of means = 0.9952452; Min. Cor. of means = 0.9917264; Max. Cor. of means = 0.9981713
## Warning in mean.default(res$PercDiff_sd_mean): argument is not numeric or
## logical: returning NA## Warning in min(res$PercDiff_sd_mean): no non-missing arguments to min; returning
## Inf## Warning in max(res$PercDiff_sd_mean): no non-missing arguments to max; returning
## -InfMedian mean %diff of SD = ; Mean mean %diff of SD = NA; Min. mean %diff of SD = Inf; Max. mean %diff of SD = -Inf
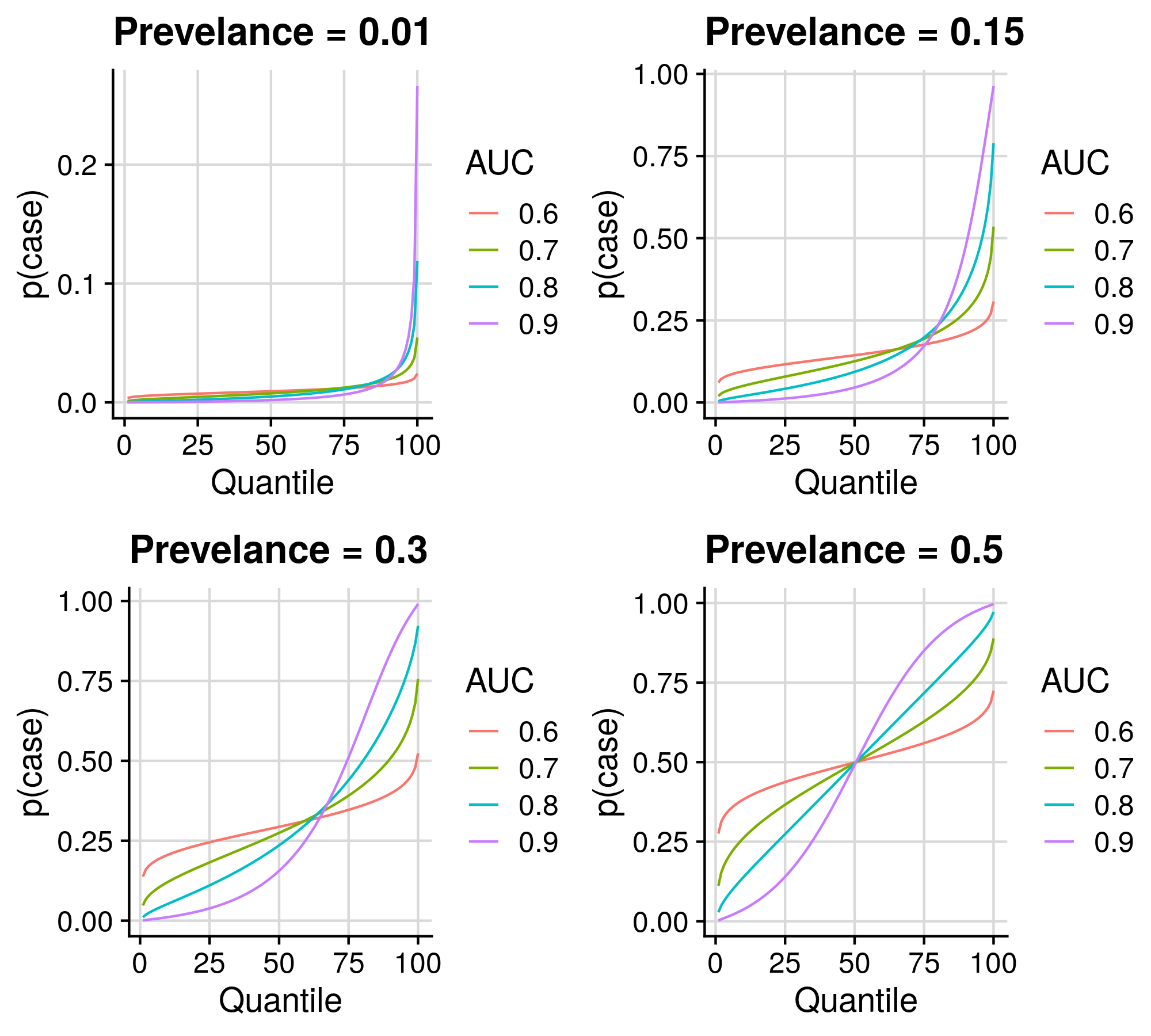
1.3 Demonstrate potential of polygenic scores
Here we will create a series of plots showing estimates on the absolute scale across PRS quantiles given a range of phenotype distribution and PRS AUC/R2.
1.3.1 Binary outcomes
Show code
# Thank you for Alex Gillet for her work developing this code.
ccprobs.f <- function(PRS_auc=0.641, prev=0.7463, n_quantile=20){
# Convert AUC into cohen's d
d <- sqrt(2)*qnorm(PRS_auc)
# Set mean difference between cases and control polygenic scores
mu_case <- d
mu_control <- 0
# Estimate mean and variance of polygenic scores across case and control
varPRS <- prev*(1+(d^2) - (d*prev)^2) + (1-prev)*(1 - (d*prev)^2)
E_PRS <- d*prev
# Estimate polygenic score quantiles
by_quant<-1/n_quantile
p_quant <- seq(by_quant, 1-by_quant, by=by_quant)
quant_vals_PRS <- rep(0, length(p_quant))
quant_f_solve <- function(x, prev, d, pq){prev*pnorm(x-d) + (1-prev)*pnorm(x) - pq}
for(i in 1:length(p_quant)){
quant_vals_PRS[i] <- unlist(uniroot(quant_f_solve, prev=prev, d=d, pq= p_quant[i], interval=c(-2.5, 2.5), extendInt = "yes", tol=6e-12)$root)
}
# Create a table for output
ul_qv_PRS <- matrix(0, ncol=2, nrow=n_quantile)
ul_qv_PRS[1,1] <- -Inf
ul_qv_PRS[2:n_quantile,1] <- quant_vals_PRS
ul_qv_PRS[1:(n_quantile-1),2] <- quant_vals_PRS
ul_qv_PRS[n_quantile,2] <- Inf
ul_qv_PRS<-cbind(ul_qv_PRS, (ul_qv_PRS[,1:2]-E_PRS)/sqrt(varPRS))
# Estimate case control proportion for each quantile
prob_quantile_case <- pnorm(ul_qv_PRS[,2], mean = mu_case) - pnorm(ul_qv_PRS[,1], mean = mu_case)
prob_quantile_control <- pnorm(ul_qv_PRS[,2], mean = mu_control) - pnorm(ul_qv_PRS[,1], mean = mu_control)
p_case_quantile <- (prob_quantile_case*prev)/by_quant
p_cont_quantile <- (prob_quantile_control*(1-prev))/by_quant
# Estimate OR comparing each quantile to bottom quantile
OR <- p_case_quantile/p_cont_quantile
OR <- OR/OR[1]
# Return output
out <- cbind(ul_qv_PRS[,3:4],p_cont_quantile, p_case_quantile, OR)
row.names(out) <- 1:n_quantile
colnames(out) <- c("q_min", "q_max","p_control", "p_case", "OR")
data.frame(out)
}
k<-as.character(c(0.01,0.15,0.3,0.5))
auc<-as.character(seq(0.6, 0.9, by=0.1))
library(ggplot2)
library(cowplot)
plot_list<-list()
plot_list_OR<-list()
res_all<-NULL
for(i in k){
res_i<-NULL
for(j in auc){
res_i_j<-ccprobs.f(PRS_auc=as.numeric(j), prev=as.numeric(i), n_quantile=100)
res_i_j$Quantile<-1:nrow(res_i_j)
res_i_j$auc<-j
res_i_j$k<-i
res_i_j$OR<-res_i_j$p_case/as.numeric(i)
res_i<-rbind(res_i, res_i_j)
}
res_all<-rbind(res_all, res_i)
plot_list[[i]]<-ggplot(res_i, aes(x=Quantile, y=p_case, group=auc, colour=auc)) +
geom_line() +
labs(y="p(case)", title=paste0('Prevelance = ',i), colour='AUC') +
theme_half_open() +
background_grid()
plot_list_OR[[i]]<-ggplot(res_i, aes(x=Quantile, y=OR, group=auc, colour=auc)) +
geom_line() +
labs(y="OR", title=paste0('Prevelance = ',i), colour='AUC') +
theme_half_open() +
background_grid()
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Binary_sim.png'), units='px', res=300, width=2000, height=1800)
plot_grid(plotlist=plot_list, ncol = 2)
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Binary_sim_OR.png'), units='px', res=300, width=2000, height=1800)
plot_grid(plotlist=plot_list_OR, ncol = 2)
dev.off()
write.csv(res_all, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Binary_sim.csv', row.names=F, quote=F)Show results
| q_min | q_max | p_control | p_case | OR | Quantile | auc | k |
|---|---|---|---|---|---|---|---|
| -Inf | -2.326 | 0.996 | 0.004 | 0.366 | 1 | 0.6 | 0.01 |
| -2.326 | -2.054 | 0.996 | 0.004 | 0.433 | 2 | 0.6 | 0.01 |
| -2.054 | -1.881 | 0.995 | 0.005 | 0.467 | 3 | 0.6 | 0.01 |
| -1.881 | -1.751 | 0.995 | 0.005 | 0.493 | 4 | 0.6 | 0.01 |
| -1.751 | -1.645 | 0.995 | 0.005 | 0.514 | 5 | 0.6 | 0.01 |
| -1.645 | -1.555 | 0.995 | 0.005 | 0.532 | 6 | 0.6 | 0.01 |
| -1.555 | -1.476 | 0.995 | 0.005 | 0.548 | 7 | 0.6 | 0.01 |
| -1.476 | -1.405 | 0.994 | 0.006 | 0.563 | 8 | 0.6 | 0.01 |
| -1.405 | -1.341 | 0.994 | 0.006 | 0.577 | 9 | 0.6 | 0.01 |
| -1.341 | -1.281 | 0.994 | 0.006 | 0.589 | 10 | 0.6 | 0.01 |
| -1.281 | -1.226 | 0.994 | 0.006 | 0.602 | 11 | 0.6 | 0.01 |
| -1.226 | -1.175 | 0.994 | 0.006 | 0.613 | 12 | 0.6 | 0.01 |
| -1.175 | -1.126 | 0.994 | 0.006 | 0.624 | 13 | 0.6 | 0.01 |
| -1.126 | -1.080 | 0.994 | 0.006 | 0.635 | 14 | 0.6 | 0.01 |
| -1.080 | -1.036 | 0.994 | 0.006 | 0.645 | 15 | 0.6 | 0.01 |
| -1.036 | -0.994 | 0.993 | 0.007 | 0.655 | 16 | 0.6 | 0.01 |
| -0.994 | -0.954 | 0.993 | 0.007 | 0.664 | 17 | 0.6 | 0.01 |
| -0.954 | -0.915 | 0.993 | 0.007 | 0.674 | 18 | 0.6 | 0.01 |
| -0.915 | -0.878 | 0.993 | 0.007 | 0.683 | 19 | 0.6 | 0.01 |
| -0.878 | -0.842 | 0.993 | 0.007 | 0.692 | 20 | 0.6 | 0.01 |
| -0.842 | -0.806 | 0.993 | 0.007 | 0.701 | 21 | 0.6 | 0.01 |
| -0.806 | -0.772 | 0.993 | 0.007 | 0.710 | 22 | 0.6 | 0.01 |
| -0.772 | -0.739 | 0.993 | 0.007 | 0.718 | 23 | 0.6 | 0.01 |
| -0.739 | -0.706 | 0.993 | 0.007 | 0.727 | 24 | 0.6 | 0.01 |
| -0.706 | -0.675 | 0.993 | 0.007 | 0.735 | 25 | 0.6 | 0.01 |
| -0.675 | -0.643 | 0.993 | 0.007 | 0.743 | 26 | 0.6 | 0.01 |
| -0.643 | -0.613 | 0.992 | 0.008 | 0.752 | 27 | 0.6 | 0.01 |
| -0.613 | -0.583 | 0.992 | 0.008 | 0.760 | 28 | 0.6 | 0.01 |
| -0.583 | -0.553 | 0.992 | 0.008 | 0.768 | 29 | 0.6 | 0.01 |
| -0.553 | -0.524 | 0.992 | 0.008 | 0.776 | 30 | 0.6 | 0.01 |
| -0.524 | -0.496 | 0.992 | 0.008 | 0.784 | 31 | 0.6 | 0.01 |
| -0.496 | -0.468 | 0.992 | 0.008 | 0.792 | 32 | 0.6 | 0.01 |
| -0.468 | -0.440 | 0.992 | 0.008 | 0.800 | 33 | 0.6 | 0.01 |
| -0.440 | -0.413 | 0.992 | 0.008 | 0.808 | 34 | 0.6 | 0.01 |
| -0.413 | -0.385 | 0.992 | 0.008 | 0.815 | 35 | 0.6 | 0.01 |
| -0.385 | -0.359 | 0.992 | 0.008 | 0.823 | 36 | 0.6 | 0.01 |
| -0.359 | -0.332 | 0.992 | 0.008 | 0.831 | 37 | 0.6 | 0.01 |
| -0.332 | -0.306 | 0.992 | 0.008 | 0.839 | 38 | 0.6 | 0.01 |
| -0.306 | -0.279 | 0.992 | 0.008 | 0.847 | 39 | 0.6 | 0.01 |
| -0.279 | -0.253 | 0.991 | 0.009 | 0.855 | 40 | 0.6 | 0.01 |
| -0.253 | -0.228 | 0.991 | 0.009 | 0.863 | 41 | 0.6 | 0.01 |
| -0.228 | -0.202 | 0.991 | 0.009 | 0.871 | 42 | 0.6 | 0.01 |
| -0.202 | -0.176 | 0.991 | 0.009 | 0.879 | 43 | 0.6 | 0.01 |
| -0.176 | -0.151 | 0.991 | 0.009 | 0.887 | 44 | 0.6 | 0.01 |
| -0.151 | -0.126 | 0.991 | 0.009 | 0.895 | 45 | 0.6 | 0.01 |
| -0.126 | -0.101 | 0.991 | 0.009 | 0.903 | 46 | 0.6 | 0.01 |
| -0.101 | -0.075 | 0.991 | 0.009 | 0.911 | 47 | 0.6 | 0.01 |
| -0.075 | -0.050 | 0.991 | 0.009 | 0.919 | 48 | 0.6 | 0.01 |
| -0.050 | -0.025 | 0.991 | 0.009 | 0.927 | 49 | 0.6 | 0.01 |
| -0.025 | 0.000 | 0.991 | 0.009 | 0.935 | 50 | 0.6 | 0.01 |
| 0.000 | 0.025 | 0.991 | 0.009 | 0.944 | 51 | 0.6 | 0.01 |
| 0.025 | 0.050 | 0.990 | 0.010 | 0.952 | 52 | 0.6 | 0.01 |
| 0.050 | 0.075 | 0.990 | 0.010 | 0.961 | 53 | 0.6 | 0.01 |
| 0.075 | 0.100 | 0.990 | 0.010 | 0.969 | 54 | 0.6 | 0.01 |
| 0.100 | 0.126 | 0.990 | 0.010 | 0.978 | 55 | 0.6 | 0.01 |
| 0.126 | 0.151 | 0.990 | 0.010 | 0.987 | 56 | 0.6 | 0.01 |
| 0.151 | 0.176 | 0.990 | 0.010 | 0.996 | 57 | 0.6 | 0.01 |
| 0.176 | 0.202 | 0.990 | 0.010 | 1.005 | 58 | 0.6 | 0.01 |
| 0.202 | 0.227 | 0.990 | 0.010 | 1.014 | 59 | 0.6 | 0.01 |
| 0.227 | 0.253 | 0.990 | 0.010 | 1.023 | 60 | 0.6 | 0.01 |
| 0.253 | 0.279 | 0.990 | 0.010 | 1.033 | 61 | 0.6 | 0.01 |
| 0.279 | 0.305 | 0.990 | 0.010 | 1.042 | 62 | 0.6 | 0.01 |
| 0.305 | 0.332 | 0.989 | 0.011 | 1.052 | 63 | 0.6 | 0.01 |
| 0.332 | 0.358 | 0.989 | 0.011 | 1.062 | 64 | 0.6 | 0.01 |
| 0.358 | 0.385 | 0.989 | 0.011 | 1.072 | 65 | 0.6 | 0.01 |
| 0.385 | 0.412 | 0.989 | 0.011 | 1.082 | 66 | 0.6 | 0.01 |
| 0.412 | 0.440 | 0.989 | 0.011 | 1.093 | 67 | 0.6 | 0.01 |
| 0.440 | 0.468 | 0.989 | 0.011 | 1.104 | 68 | 0.6 | 0.01 |
| 0.468 | 0.496 | 0.989 | 0.011 | 1.115 | 69 | 0.6 | 0.01 |
| 0.496 | 0.524 | 0.989 | 0.011 | 1.126 | 70 | 0.6 | 0.01 |
| 0.524 | 0.553 | 0.989 | 0.011 | 1.138 | 71 | 0.6 | 0.01 |
| 0.553 | 0.583 | 0.989 | 0.011 | 1.149 | 72 | 0.6 | 0.01 |
| 0.583 | 0.613 | 0.988 | 0.012 | 1.162 | 73 | 0.6 | 0.01 |
| 0.613 | 0.643 | 0.988 | 0.012 | 1.174 | 74 | 0.6 | 0.01 |
| 0.643 | 0.674 | 0.988 | 0.012 | 1.187 | 75 | 0.6 | 0.01 |
| 0.674 | 0.706 | 0.988 | 0.012 | 1.200 | 76 | 0.6 | 0.01 |
| 0.706 | 0.739 | 0.988 | 0.012 | 1.214 | 77 | 0.6 | 0.01 |
| 0.739 | 0.772 | 0.988 | 0.012 | 1.228 | 78 | 0.6 | 0.01 |
| 0.772 | 0.806 | 0.988 | 0.012 | 1.243 | 79 | 0.6 | 0.01 |
| 0.806 | 0.842 | 0.987 | 0.013 | 1.258 | 80 | 0.6 | 0.01 |
| 0.842 | 0.878 | 0.987 | 0.013 | 1.274 | 81 | 0.6 | 0.01 |
| 0.878 | 0.915 | 0.987 | 0.013 | 1.291 | 82 | 0.6 | 0.01 |
| 0.915 | 0.954 | 0.987 | 0.013 | 1.309 | 83 | 0.6 | 0.01 |
| 0.954 | 0.994 | 0.987 | 0.013 | 1.327 | 84 | 0.6 | 0.01 |
| 0.994 | 1.036 | 0.987 | 0.013 | 1.347 | 85 | 0.6 | 0.01 |
| 1.036 | 1.080 | 0.986 | 0.014 | 1.367 | 86 | 0.6 | 0.01 |
| 1.080 | 1.126 | 0.986 | 0.014 | 1.389 | 87 | 0.6 | 0.01 |
| 1.126 | 1.175 | 0.986 | 0.014 | 1.413 | 88 | 0.6 | 0.01 |
| 1.175 | 1.227 | 0.986 | 0.014 | 1.438 | 89 | 0.6 | 0.01 |
| 1.227 | 1.282 | 0.985 | 0.015 | 1.465 | 90 | 0.6 | 0.01 |
| 1.282 | 1.341 | 0.985 | 0.015 | 1.495 | 91 | 0.6 | 0.01 |
| 1.341 | 1.405 | 0.985 | 0.015 | 1.528 | 92 | 0.6 | 0.01 |
| 1.405 | 1.476 | 0.984 | 0.016 | 1.565 | 93 | 0.6 | 0.01 |
| 1.476 | 1.555 | 0.984 | 0.016 | 1.606 | 94 | 0.6 | 0.01 |
| 1.555 | 1.645 | 0.983 | 0.017 | 1.655 | 95 | 0.6 | 0.01 |
| 1.645 | 1.751 | 0.983 | 0.017 | 1.713 | 96 | 0.6 | 0.01 |
| 1.751 | 1.881 | 0.982 | 0.018 | 1.785 | 97 | 0.6 | 0.01 |
| 1.881 | 2.054 | 0.981 | 0.019 | 1.881 | 98 | 0.6 | 0.01 |
| 2.054 | 2.327 | 0.980 | 0.020 | 2.029 | 99 | 0.6 | 0.01 |
| 2.327 | Inf | 0.976 | 0.024 | 2.422 | 100 | 0.6 | 0.01 |
| -Inf | -2.324 | 0.999 | 0.001 | 0.109 | 1 | 0.7 | 0.01 |
| -2.324 | -2.052 | 0.998 | 0.002 | 0.153 | 2 | 0.7 | 0.01 |
| -2.052 | -1.879 | 0.998 | 0.002 | 0.179 | 3 | 0.7 | 0.01 |
| -1.879 | -1.749 | 0.998 | 0.002 | 0.200 | 4 | 0.7 | 0.01 |
| -1.749 | -1.644 | 0.998 | 0.002 | 0.218 | 5 | 0.7 | 0.01 |
| -1.644 | -1.554 | 0.998 | 0.002 | 0.235 | 6 | 0.7 | 0.01 |
| -1.554 | -1.475 | 0.998 | 0.002 | 0.250 | 7 | 0.7 | 0.01 |
| -1.475 | -1.404 | 0.997 | 0.003 | 0.264 | 8 | 0.7 | 0.01 |
| -1.404 | -1.340 | 0.997 | 0.003 | 0.277 | 9 | 0.7 | 0.01 |
| -1.340 | -1.281 | 0.997 | 0.003 | 0.290 | 10 | 0.7 | 0.01 |
| -1.281 | -1.226 | 0.997 | 0.003 | 0.303 | 11 | 0.7 | 0.01 |
| -1.226 | -1.175 | 0.997 | 0.003 | 0.315 | 12 | 0.7 | 0.01 |
| -1.175 | -1.126 | 0.997 | 0.003 | 0.327 | 13 | 0.7 | 0.01 |
| -1.126 | -1.080 | 0.997 | 0.003 | 0.339 | 14 | 0.7 | 0.01 |
| -1.080 | -1.036 | 0.996 | 0.004 | 0.350 | 15 | 0.7 | 0.01 |
| -1.036 | -0.994 | 0.996 | 0.004 | 0.361 | 16 | 0.7 | 0.01 |
| -0.994 | -0.954 | 0.996 | 0.004 | 0.373 | 17 | 0.7 | 0.01 |
| -0.954 | -0.915 | 0.996 | 0.004 | 0.384 | 18 | 0.7 | 0.01 |
| -0.915 | -0.878 | 0.996 | 0.004 | 0.395 | 19 | 0.7 | 0.01 |
| -0.878 | -0.842 | 0.996 | 0.004 | 0.405 | 20 | 0.7 | 0.01 |
| -0.842 | -0.806 | 0.996 | 0.004 | 0.416 | 21 | 0.7 | 0.01 |
| -0.806 | -0.772 | 0.996 | 0.004 | 0.427 | 22 | 0.7 | 0.01 |
| -0.772 | -0.739 | 0.996 | 0.004 | 0.438 | 23 | 0.7 | 0.01 |
| -0.739 | -0.706 | 0.996 | 0.004 | 0.449 | 24 | 0.7 | 0.01 |
| -0.706 | -0.675 | 0.995 | 0.005 | 0.460 | 25 | 0.7 | 0.01 |
| -0.675 | -0.644 | 0.995 | 0.005 | 0.470 | 26 | 0.7 | 0.01 |
| -0.644 | -0.613 | 0.995 | 0.005 | 0.481 | 27 | 0.7 | 0.01 |
| -0.613 | -0.583 | 0.995 | 0.005 | 0.492 | 28 | 0.7 | 0.01 |
| -0.583 | -0.554 | 0.995 | 0.005 | 0.503 | 29 | 0.7 | 0.01 |
| -0.554 | -0.525 | 0.995 | 0.005 | 0.514 | 30 | 0.7 | 0.01 |
| -0.525 | -0.496 | 0.995 | 0.005 | 0.525 | 31 | 0.7 | 0.01 |
| -0.496 | -0.468 | 0.995 | 0.005 | 0.536 | 32 | 0.7 | 0.01 |
| -0.468 | -0.440 | 0.995 | 0.005 | 0.547 | 33 | 0.7 | 0.01 |
| -0.440 | -0.413 | 0.994 | 0.006 | 0.559 | 34 | 0.7 | 0.01 |
| -0.413 | -0.386 | 0.994 | 0.006 | 0.570 | 35 | 0.7 | 0.01 |
| -0.386 | -0.359 | 0.994 | 0.006 | 0.582 | 36 | 0.7 | 0.01 |
| -0.359 | -0.332 | 0.994 | 0.006 | 0.593 | 37 | 0.7 | 0.01 |
| -0.332 | -0.306 | 0.994 | 0.006 | 0.605 | 38 | 0.7 | 0.01 |
| -0.306 | -0.280 | 0.994 | 0.006 | 0.617 | 39 | 0.7 | 0.01 |
| -0.280 | -0.254 | 0.994 | 0.006 | 0.629 | 40 | 0.7 | 0.01 |
| -0.254 | -0.228 | 0.994 | 0.006 | 0.641 | 41 | 0.7 | 0.01 |
| -0.228 | -0.202 | 0.993 | 0.007 | 0.653 | 42 | 0.7 | 0.01 |
| -0.202 | -0.177 | 0.993 | 0.007 | 0.666 | 43 | 0.7 | 0.01 |
| -0.177 | -0.152 | 0.993 | 0.007 | 0.678 | 44 | 0.7 | 0.01 |
| -0.152 | -0.126 | 0.993 | 0.007 | 0.691 | 45 | 0.7 | 0.01 |
| -0.126 | -0.101 | 0.993 | 0.007 | 0.704 | 46 | 0.7 | 0.01 |
| -0.101 | -0.076 | 0.993 | 0.007 | 0.717 | 47 | 0.7 | 0.01 |
| -0.076 | -0.051 | 0.993 | 0.007 | 0.731 | 48 | 0.7 | 0.01 |
| -0.051 | -0.026 | 0.993 | 0.007 | 0.744 | 49 | 0.7 | 0.01 |
| -0.026 | -0.001 | 0.992 | 0.008 | 0.758 | 50 | 0.7 | 0.01 |
| -0.001 | 0.024 | 0.992 | 0.008 | 0.772 | 51 | 0.7 | 0.01 |
| 0.024 | 0.050 | 0.992 | 0.008 | 0.787 | 52 | 0.7 | 0.01 |
| 0.050 | 0.075 | 0.992 | 0.008 | 0.801 | 53 | 0.7 | 0.01 |
| 0.075 | 0.100 | 0.992 | 0.008 | 0.816 | 54 | 0.7 | 0.01 |
| 0.100 | 0.125 | 0.992 | 0.008 | 0.832 | 55 | 0.7 | 0.01 |
| 0.125 | 0.150 | 0.992 | 0.008 | 0.847 | 56 | 0.7 | 0.01 |
| 0.150 | 0.176 | 0.991 | 0.009 | 0.863 | 57 | 0.7 | 0.01 |
| 0.176 | 0.201 | 0.991 | 0.009 | 0.880 | 58 | 0.7 | 0.01 |
| 0.201 | 0.227 | 0.991 | 0.009 | 0.896 | 59 | 0.7 | 0.01 |
| 0.227 | 0.253 | 0.991 | 0.009 | 0.914 | 60 | 0.7 | 0.01 |
| 0.253 | 0.279 | 0.991 | 0.009 | 0.931 | 61 | 0.7 | 0.01 |
| 0.279 | 0.305 | 0.991 | 0.009 | 0.949 | 62 | 0.7 | 0.01 |
| 0.305 | 0.331 | 0.990 | 0.010 | 0.968 | 63 | 0.7 | 0.01 |
| 0.331 | 0.358 | 0.990 | 0.010 | 0.987 | 64 | 0.7 | 0.01 |
| 0.358 | 0.385 | 0.990 | 0.010 | 1.007 | 65 | 0.7 | 0.01 |
| 0.385 | 0.412 | 0.990 | 0.010 | 1.027 | 66 | 0.7 | 0.01 |
| 0.412 | 0.439 | 0.990 | 0.010 | 1.048 | 67 | 0.7 | 0.01 |
| 0.439 | 0.467 | 0.989 | 0.011 | 1.069 | 68 | 0.7 | 0.01 |
| 0.467 | 0.495 | 0.989 | 0.011 | 1.091 | 69 | 0.7 | 0.01 |
| 0.495 | 0.524 | 0.989 | 0.011 | 1.114 | 70 | 0.7 | 0.01 |
| 0.524 | 0.553 | 0.989 | 0.011 | 1.138 | 71 | 0.7 | 0.01 |
| 0.553 | 0.582 | 0.988 | 0.012 | 1.163 | 72 | 0.7 | 0.01 |
| 0.582 | 0.612 | 0.988 | 0.012 | 1.189 | 73 | 0.7 | 0.01 |
| 0.612 | 0.643 | 0.988 | 0.012 | 1.215 | 74 | 0.7 | 0.01 |
| 0.643 | 0.674 | 0.988 | 0.012 | 1.243 | 75 | 0.7 | 0.01 |
| 0.674 | 0.706 | 0.987 | 0.013 | 1.272 | 76 | 0.7 | 0.01 |
| 0.706 | 0.738 | 0.987 | 0.013 | 1.303 | 77 | 0.7 | 0.01 |
| 0.738 | 0.772 | 0.987 | 0.013 | 1.335 | 78 | 0.7 | 0.01 |
| 0.772 | 0.806 | 0.986 | 0.014 | 1.368 | 79 | 0.7 | 0.01 |
| 0.806 | 0.841 | 0.986 | 0.014 | 1.403 | 80 | 0.7 | 0.01 |
| 0.841 | 0.878 | 0.986 | 0.014 | 1.441 | 81 | 0.7 | 0.01 |
| 0.878 | 0.915 | 0.985 | 0.015 | 1.480 | 82 | 0.7 | 0.01 |
| 0.915 | 0.954 | 0.985 | 0.015 | 1.522 | 83 | 0.7 | 0.01 |
| 0.954 | 0.994 | 0.984 | 0.016 | 1.567 | 84 | 0.7 | 0.01 |
| 0.994 | 1.036 | 0.984 | 0.016 | 1.615 | 85 | 0.7 | 0.01 |
| 1.036 | 1.080 | 0.983 | 0.017 | 1.666 | 86 | 0.7 | 0.01 |
| 1.080 | 1.126 | 0.983 | 0.017 | 1.722 | 87 | 0.7 | 0.01 |
| 1.126 | 1.175 | 0.982 | 0.018 | 1.783 | 88 | 0.7 | 0.01 |
| 1.175 | 1.227 | 0.982 | 0.018 | 1.849 | 89 | 0.7 | 0.01 |
| 1.227 | 1.282 | 0.981 | 0.019 | 1.922 | 90 | 0.7 | 0.01 |
| 1.282 | 1.341 | 0.980 | 0.020 | 2.004 | 91 | 0.7 | 0.01 |
| 1.341 | 1.405 | 0.979 | 0.021 | 2.097 | 92 | 0.7 | 0.01 |
| 1.405 | 1.476 | 0.978 | 0.022 | 2.202 | 93 | 0.7 | 0.01 |
| 1.476 | 1.556 | 0.977 | 0.023 | 2.325 | 94 | 0.7 | 0.01 |
| 1.556 | 1.646 | 0.975 | 0.025 | 2.472 | 95 | 0.7 | 0.01 |
| 1.646 | 1.752 | 0.973 | 0.027 | 2.654 | 96 | 0.7 | 0.01 |
| 1.752 | 1.882 | 0.971 | 0.029 | 2.889 | 97 | 0.7 | 0.01 |
| 1.882 | 2.056 | 0.968 | 0.032 | 3.219 | 98 | 0.7 | 0.01 |
| 2.056 | 2.330 | 0.962 | 0.038 | 3.760 | 99 | 0.7 | 0.01 |
| 2.330 | Inf | 0.945 | 0.055 | 5.458 | 100 | 0.7 | 0.01 |
| -Inf | -2.318 | 1.000 | 0.000 | 0.022 | 1 | 0.8 | 0.01 |
| -2.318 | -2.047 | 1.000 | 0.000 | 0.038 | 2 | 0.8 | 0.01 |
| -2.047 | -1.875 | 1.000 | 0.000 | 0.048 | 3 | 0.8 | 0.01 |
| -1.875 | -1.746 | 0.999 | 0.001 | 0.058 | 4 | 0.8 | 0.01 |
| -1.746 | -1.641 | 0.999 | 0.001 | 0.066 | 5 | 0.8 | 0.01 |
| -1.641 | -1.551 | 0.999 | 0.001 | 0.075 | 6 | 0.8 | 0.01 |
| -1.551 | -1.472 | 0.999 | 0.001 | 0.082 | 7 | 0.8 | 0.01 |
| -1.472 | -1.402 | 0.999 | 0.001 | 0.090 | 8 | 0.8 | 0.01 |
| -1.402 | -1.338 | 0.999 | 0.001 | 0.098 | 9 | 0.8 | 0.01 |
| -1.338 | -1.279 | 0.999 | 0.001 | 0.105 | 10 | 0.8 | 0.01 |
| -1.279 | -1.224 | 0.999 | 0.001 | 0.112 | 11 | 0.8 | 0.01 |
| -1.224 | -1.173 | 0.999 | 0.001 | 0.120 | 12 | 0.8 | 0.01 |
| -1.173 | -1.125 | 0.999 | 0.001 | 0.127 | 13 | 0.8 | 0.01 |
| -1.125 | -1.079 | 0.999 | 0.001 | 0.135 | 14 | 0.8 | 0.01 |
| -1.079 | -1.035 | 0.999 | 0.001 | 0.142 | 15 | 0.8 | 0.01 |
| -1.035 | -0.993 | 0.999 | 0.001 | 0.149 | 16 | 0.8 | 0.01 |
| -0.993 | -0.953 | 0.998 | 0.002 | 0.157 | 17 | 0.8 | 0.01 |
| -0.953 | -0.915 | 0.998 | 0.002 | 0.164 | 18 | 0.8 | 0.01 |
| -0.915 | -0.877 | 0.998 | 0.002 | 0.172 | 19 | 0.8 | 0.01 |
| -0.877 | -0.841 | 0.998 | 0.002 | 0.180 | 20 | 0.8 | 0.01 |
| -0.841 | -0.806 | 0.998 | 0.002 | 0.188 | 21 | 0.8 | 0.01 |
| -0.806 | -0.772 | 0.998 | 0.002 | 0.196 | 22 | 0.8 | 0.01 |
| -0.772 | -0.739 | 0.998 | 0.002 | 0.204 | 23 | 0.8 | 0.01 |
| -0.739 | -0.706 | 0.998 | 0.002 | 0.212 | 24 | 0.8 | 0.01 |
| -0.706 | -0.675 | 0.998 | 0.002 | 0.220 | 25 | 0.8 | 0.01 |
| -0.675 | -0.644 | 0.998 | 0.002 | 0.228 | 26 | 0.8 | 0.01 |
| -0.644 | -0.613 | 0.998 | 0.002 | 0.237 | 27 | 0.8 | 0.01 |
| -0.613 | -0.583 | 0.998 | 0.002 | 0.246 | 28 | 0.8 | 0.01 |
| -0.583 | -0.554 | 0.997 | 0.003 | 0.255 | 29 | 0.8 | 0.01 |
| -0.554 | -0.525 | 0.997 | 0.003 | 0.264 | 30 | 0.8 | 0.01 |
| -0.525 | -0.497 | 0.997 | 0.003 | 0.273 | 31 | 0.8 | 0.01 |
| -0.497 | -0.469 | 0.997 | 0.003 | 0.282 | 32 | 0.8 | 0.01 |
| -0.469 | -0.441 | 0.997 | 0.003 | 0.292 | 33 | 0.8 | 0.01 |
| -0.441 | -0.414 | 0.997 | 0.003 | 0.301 | 34 | 0.8 | 0.01 |
| -0.414 | -0.387 | 0.997 | 0.003 | 0.311 | 35 | 0.8 | 0.01 |
| -0.387 | -0.360 | 0.997 | 0.003 | 0.322 | 36 | 0.8 | 0.01 |
| -0.360 | -0.333 | 0.997 | 0.003 | 0.332 | 37 | 0.8 | 0.01 |
| -0.333 | -0.307 | 0.997 | 0.003 | 0.343 | 38 | 0.8 | 0.01 |
| -0.307 | -0.281 | 0.996 | 0.004 | 0.353 | 39 | 0.8 | 0.01 |
| -0.281 | -0.255 | 0.996 | 0.004 | 0.365 | 40 | 0.8 | 0.01 |
| -0.255 | -0.229 | 0.996 | 0.004 | 0.376 | 41 | 0.8 | 0.01 |
| -0.229 | -0.204 | 0.996 | 0.004 | 0.388 | 42 | 0.8 | 0.01 |
| -0.204 | -0.178 | 0.996 | 0.004 | 0.400 | 43 | 0.8 | 0.01 |
| -0.178 | -0.153 | 0.996 | 0.004 | 0.412 | 44 | 0.8 | 0.01 |
| -0.153 | -0.128 | 0.996 | 0.004 | 0.425 | 45 | 0.8 | 0.01 |
| -0.128 | -0.102 | 0.996 | 0.004 | 0.438 | 46 | 0.8 | 0.01 |
| -0.102 | -0.077 | 0.995 | 0.005 | 0.451 | 47 | 0.8 | 0.01 |
| -0.077 | -0.052 | 0.995 | 0.005 | 0.465 | 48 | 0.8 | 0.01 |
| -0.052 | -0.027 | 0.995 | 0.005 | 0.479 | 49 | 0.8 | 0.01 |
| -0.027 | -0.002 | 0.995 | 0.005 | 0.493 | 50 | 0.8 | 0.01 |
| -0.002 | 0.023 | 0.995 | 0.005 | 0.508 | 51 | 0.8 | 0.01 |
| 0.023 | 0.048 | 0.995 | 0.005 | 0.524 | 52 | 0.8 | 0.01 |
| 0.048 | 0.073 | 0.995 | 0.005 | 0.539 | 53 | 0.8 | 0.01 |
| 0.073 | 0.098 | 0.994 | 0.006 | 0.556 | 54 | 0.8 | 0.01 |
| 0.098 | 0.123 | 0.994 | 0.006 | 0.573 | 55 | 0.8 | 0.01 |
| 0.123 | 0.148 | 0.994 | 0.006 | 0.590 | 56 | 0.8 | 0.01 |
| 0.148 | 0.174 | 0.994 | 0.006 | 0.608 | 57 | 0.8 | 0.01 |
| 0.174 | 0.199 | 0.994 | 0.006 | 0.627 | 58 | 0.8 | 0.01 |
| 0.199 | 0.225 | 0.994 | 0.006 | 0.646 | 59 | 0.8 | 0.01 |
| 0.225 | 0.251 | 0.993 | 0.007 | 0.667 | 60 | 0.8 | 0.01 |
| 0.251 | 0.277 | 0.993 | 0.007 | 0.687 | 61 | 0.8 | 0.01 |
| 0.277 | 0.303 | 0.993 | 0.007 | 0.709 | 62 | 0.8 | 0.01 |
| 0.303 | 0.329 | 0.993 | 0.007 | 0.732 | 63 | 0.8 | 0.01 |
| 0.329 | 0.356 | 0.992 | 0.008 | 0.755 | 64 | 0.8 | 0.01 |
| 0.356 | 0.383 | 0.992 | 0.008 | 0.779 | 65 | 0.8 | 0.01 |
| 0.383 | 0.410 | 0.992 | 0.008 | 0.805 | 66 | 0.8 | 0.01 |
| 0.410 | 0.437 | 0.992 | 0.008 | 0.831 | 67 | 0.8 | 0.01 |
| 0.437 | 0.465 | 0.991 | 0.009 | 0.859 | 68 | 0.8 | 0.01 |
| 0.465 | 0.493 | 0.991 | 0.009 | 0.888 | 69 | 0.8 | 0.01 |
| 0.493 | 0.522 | 0.991 | 0.009 | 0.918 | 70 | 0.8 | 0.01 |
| 0.522 | 0.551 | 0.990 | 0.010 | 0.950 | 71 | 0.8 | 0.01 |
| 0.551 | 0.580 | 0.990 | 0.010 | 0.984 | 72 | 0.8 | 0.01 |
| 0.580 | 0.610 | 0.990 | 0.010 | 1.019 | 73 | 0.8 | 0.01 |
| 0.610 | 0.641 | 0.989 | 0.011 | 1.057 | 74 | 0.8 | 0.01 |
| 0.641 | 0.672 | 0.989 | 0.011 | 1.096 | 75 | 0.8 | 0.01 |
| 0.672 | 0.704 | 0.989 | 0.011 | 1.138 | 76 | 0.8 | 0.01 |
| 0.704 | 0.736 | 0.988 | 0.012 | 1.182 | 77 | 0.8 | 0.01 |
| 0.736 | 0.770 | 0.988 | 0.012 | 1.229 | 78 | 0.8 | 0.01 |
| 0.770 | 0.804 | 0.987 | 0.013 | 1.279 | 79 | 0.8 | 0.01 |
| 0.804 | 0.839 | 0.987 | 0.013 | 1.333 | 80 | 0.8 | 0.01 |
| 0.839 | 0.876 | 0.986 | 0.014 | 1.391 | 81 | 0.8 | 0.01 |
| 0.876 | 0.913 | 0.985 | 0.015 | 1.453 | 82 | 0.8 | 0.01 |
| 0.913 | 0.952 | 0.985 | 0.015 | 1.520 | 83 | 0.8 | 0.01 |
| 0.952 | 0.993 | 0.984 | 0.016 | 1.593 | 84 | 0.8 | 0.01 |
| 0.993 | 1.035 | 0.983 | 0.017 | 1.672 | 85 | 0.8 | 0.01 |
| 1.035 | 1.079 | 0.982 | 0.018 | 1.759 | 86 | 0.8 | 0.01 |
| 1.079 | 1.125 | 0.981 | 0.019 | 1.855 | 87 | 0.8 | 0.01 |
| 1.125 | 1.174 | 0.980 | 0.020 | 1.962 | 88 | 0.8 | 0.01 |
| 1.174 | 1.226 | 0.979 | 0.021 | 2.082 | 89 | 0.8 | 0.01 |
| 1.226 | 1.281 | 0.978 | 0.022 | 2.217 | 90 | 0.8 | 0.01 |
| 1.281 | 1.341 | 0.976 | 0.024 | 2.371 | 91 | 0.8 | 0.01 |
| 1.341 | 1.406 | 0.975 | 0.025 | 2.549 | 92 | 0.8 | 0.01 |
| 1.406 | 1.477 | 0.972 | 0.028 | 2.760 | 93 | 0.8 | 0.01 |
| 1.477 | 1.557 | 0.970 | 0.030 | 3.013 | 94 | 0.8 | 0.01 |
| 1.557 | 1.648 | 0.967 | 0.033 | 3.326 | 95 | 0.8 | 0.01 |
| 1.648 | 1.755 | 0.963 | 0.037 | 3.728 | 96 | 0.8 | 0.01 |
| 1.755 | 1.887 | 0.957 | 0.043 | 4.275 | 97 | 0.8 | 0.01 |
| 1.887 | 2.063 | 0.949 | 0.051 | 5.086 | 98 | 0.8 | 0.01 |
| 2.063 | 2.342 | 0.935 | 0.065 | 6.523 | 99 | 0.8 | 0.01 |
| 2.342 | Inf | 0.881 | 0.119 | 11.908 | 100 | 0.8 | 0.01 |
| -Inf | -2.304 | 1.000 | 0.000 | 0.002 | 1 | 0.9 | 0.01 |
| -2.304 | -2.035 | 1.000 | 0.000 | 0.004 | 2 | 0.9 | 0.01 |
| -2.035 | -1.864 | 1.000 | 0.000 | 0.006 | 3 | 0.9 | 0.01 |
| -1.864 | -1.736 | 1.000 | 0.000 | 0.007 | 4 | 0.9 | 0.01 |
| -1.736 | -1.632 | 1.000 | 0.000 | 0.009 | 5 | 0.9 | 0.01 |
| -1.632 | -1.543 | 1.000 | 0.000 | 0.011 | 6 | 0.9 | 0.01 |
| -1.543 | -1.465 | 1.000 | 0.000 | 0.013 | 7 | 0.9 | 0.01 |
| -1.465 | -1.395 | 1.000 | 0.000 | 0.015 | 8 | 0.9 | 0.01 |
| -1.395 | -1.332 | 1.000 | 0.000 | 0.016 | 9 | 0.9 | 0.01 |
| -1.332 | -1.273 | 1.000 | 0.000 | 0.018 | 10 | 0.9 | 0.01 |
| -1.273 | -1.219 | 1.000 | 0.000 | 0.020 | 11 | 0.9 | 0.01 |
| -1.219 | -1.168 | 1.000 | 0.000 | 0.022 | 12 | 0.9 | 0.01 |
| -1.168 | -1.120 | 1.000 | 0.000 | 0.025 | 13 | 0.9 | 0.01 |
| -1.120 | -1.075 | 1.000 | 0.000 | 0.027 | 14 | 0.9 | 0.01 |
| -1.075 | -1.032 | 1.000 | 0.000 | 0.029 | 15 | 0.9 | 0.01 |
| -1.032 | -0.990 | 1.000 | 0.000 | 0.031 | 16 | 0.9 | 0.01 |
| -0.990 | -0.950 | 1.000 | 0.000 | 0.034 | 17 | 0.9 | 0.01 |
| -0.950 | -0.912 | 1.000 | 0.000 | 0.036 | 18 | 0.9 | 0.01 |
| -0.912 | -0.875 | 1.000 | 0.000 | 0.039 | 19 | 0.9 | 0.01 |
| -0.875 | -0.839 | 1.000 | 0.000 | 0.042 | 20 | 0.9 | 0.01 |
| -0.839 | -0.804 | 1.000 | 0.000 | 0.044 | 21 | 0.9 | 0.01 |
| -0.804 | -0.771 | 1.000 | 0.000 | 0.047 | 22 | 0.9 | 0.01 |
| -0.771 | -0.738 | 0.999 | 0.001 | 0.050 | 23 | 0.9 | 0.01 |
| -0.738 | -0.705 | 0.999 | 0.001 | 0.053 | 24 | 0.9 | 0.01 |
| -0.705 | -0.674 | 0.999 | 0.001 | 0.057 | 25 | 0.9 | 0.01 |
| -0.674 | -0.643 | 0.999 | 0.001 | 0.060 | 26 | 0.9 | 0.01 |
| -0.643 | -0.613 | 0.999 | 0.001 | 0.064 | 27 | 0.9 | 0.01 |
| -0.613 | -0.583 | 0.999 | 0.001 | 0.067 | 28 | 0.9 | 0.01 |
| -0.583 | -0.554 | 0.999 | 0.001 | 0.071 | 29 | 0.9 | 0.01 |
| -0.554 | -0.526 | 0.999 | 0.001 | 0.075 | 30 | 0.9 | 0.01 |
| -0.526 | -0.497 | 0.999 | 0.001 | 0.079 | 31 | 0.9 | 0.01 |
| -0.497 | -0.470 | 0.999 | 0.001 | 0.083 | 32 | 0.9 | 0.01 |
| -0.470 | -0.442 | 0.999 | 0.001 | 0.087 | 33 | 0.9 | 0.01 |
| -0.442 | -0.415 | 0.999 | 0.001 | 0.092 | 34 | 0.9 | 0.01 |
| -0.415 | -0.388 | 0.999 | 0.001 | 0.096 | 35 | 0.9 | 0.01 |
| -0.388 | -0.361 | 0.999 | 0.001 | 0.101 | 36 | 0.9 | 0.01 |
| -0.361 | -0.335 | 0.999 | 0.001 | 0.106 | 37 | 0.9 | 0.01 |
| -0.335 | -0.309 | 0.999 | 0.001 | 0.112 | 38 | 0.9 | 0.01 |
| -0.309 | -0.283 | 0.999 | 0.001 | 0.117 | 39 | 0.9 | 0.01 |
| -0.283 | -0.257 | 0.999 | 0.001 | 0.123 | 40 | 0.9 | 0.01 |
| -0.257 | -0.232 | 0.999 | 0.001 | 0.129 | 41 | 0.9 | 0.01 |
| -0.232 | -0.206 | 0.999 | 0.001 | 0.135 | 42 | 0.9 | 0.01 |
| -0.206 | -0.181 | 0.999 | 0.001 | 0.141 | 43 | 0.9 | 0.01 |
| -0.181 | -0.156 | 0.999 | 0.001 | 0.148 | 44 | 0.9 | 0.01 |
| -0.156 | -0.131 | 0.998 | 0.002 | 0.155 | 45 | 0.9 | 0.01 |
| -0.131 | -0.106 | 0.998 | 0.002 | 0.162 | 46 | 0.9 | 0.01 |
| -0.106 | -0.081 | 0.998 | 0.002 | 0.170 | 47 | 0.9 | 0.01 |
| -0.081 | -0.056 | 0.998 | 0.002 | 0.178 | 48 | 0.9 | 0.01 |
| -0.056 | -0.031 | 0.998 | 0.002 | 0.186 | 49 | 0.9 | 0.01 |
| -0.031 | -0.006 | 0.998 | 0.002 | 0.195 | 50 | 0.9 | 0.01 |
| -0.006 | 0.019 | 0.998 | 0.002 | 0.204 | 51 | 0.9 | 0.01 |
| 0.019 | 0.043 | 0.998 | 0.002 | 0.213 | 52 | 0.9 | 0.01 |
| 0.043 | 0.068 | 0.998 | 0.002 | 0.223 | 53 | 0.9 | 0.01 |
| 0.068 | 0.093 | 0.998 | 0.002 | 0.234 | 54 | 0.9 | 0.01 |
| 0.093 | 0.118 | 0.998 | 0.002 | 0.245 | 55 | 0.9 | 0.01 |
| 0.118 | 0.144 | 0.997 | 0.003 | 0.256 | 56 | 0.9 | 0.01 |
| 0.144 | 0.169 | 0.997 | 0.003 | 0.269 | 57 | 0.9 | 0.01 |
| 0.169 | 0.194 | 0.997 | 0.003 | 0.281 | 58 | 0.9 | 0.01 |
| 0.194 | 0.220 | 0.997 | 0.003 | 0.295 | 59 | 0.9 | 0.01 |
| 0.220 | 0.245 | 0.997 | 0.003 | 0.309 | 60 | 0.9 | 0.01 |
| 0.245 | 0.271 | 0.997 | 0.003 | 0.324 | 61 | 0.9 | 0.01 |
| 0.271 | 0.297 | 0.997 | 0.003 | 0.340 | 62 | 0.9 | 0.01 |
| 0.297 | 0.323 | 0.996 | 0.004 | 0.357 | 63 | 0.9 | 0.01 |
| 0.323 | 0.350 | 0.996 | 0.004 | 0.374 | 64 | 0.9 | 0.01 |
| 0.350 | 0.377 | 0.996 | 0.004 | 0.393 | 65 | 0.9 | 0.01 |
| 0.377 | 0.404 | 0.996 | 0.004 | 0.413 | 66 | 0.9 | 0.01 |
| 0.404 | 0.431 | 0.996 | 0.004 | 0.434 | 67 | 0.9 | 0.01 |
| 0.431 | 0.459 | 0.995 | 0.005 | 0.456 | 68 | 0.9 | 0.01 |
| 0.459 | 0.487 | 0.995 | 0.005 | 0.480 | 69 | 0.9 | 0.01 |
| 0.487 | 0.515 | 0.995 | 0.005 | 0.506 | 70 | 0.9 | 0.01 |
| 0.515 | 0.544 | 0.995 | 0.005 | 0.533 | 71 | 0.9 | 0.01 |
| 0.544 | 0.574 | 0.994 | 0.006 | 0.562 | 72 | 0.9 | 0.01 |
| 0.574 | 0.604 | 0.994 | 0.006 | 0.594 | 73 | 0.9 | 0.01 |
| 0.604 | 0.634 | 0.994 | 0.006 | 0.628 | 74 | 0.9 | 0.01 |
| 0.634 | 0.665 | 0.993 | 0.007 | 0.664 | 75 | 0.9 | 0.01 |
| 0.665 | 0.697 | 0.993 | 0.007 | 0.703 | 76 | 0.9 | 0.01 |
| 0.697 | 0.730 | 0.993 | 0.007 | 0.746 | 77 | 0.9 | 0.01 |
| 0.730 | 0.763 | 0.992 | 0.008 | 0.793 | 78 | 0.9 | 0.01 |
| 0.763 | 0.798 | 0.992 | 0.008 | 0.843 | 79 | 0.9 | 0.01 |
| 0.798 | 0.833 | 0.991 | 0.009 | 0.898 | 80 | 0.9 | 0.01 |
| 0.833 | 0.869 | 0.990 | 0.010 | 0.959 | 81 | 0.9 | 0.01 |
| 0.869 | 0.907 | 0.990 | 0.010 | 1.026 | 82 | 0.9 | 0.01 |
| 0.907 | 0.946 | 0.989 | 0.011 | 1.101 | 83 | 0.9 | 0.01 |
| 0.946 | 0.987 | 0.988 | 0.012 | 1.183 | 84 | 0.9 | 0.01 |
| 0.987 | 1.029 | 0.987 | 0.013 | 1.276 | 85 | 0.9 | 0.01 |
| 1.029 | 1.073 | 0.986 | 0.014 | 1.381 | 86 | 0.9 | 0.01 |
| 1.073 | 1.120 | 0.985 | 0.015 | 1.499 | 87 | 0.9 | 0.01 |
| 1.120 | 1.169 | 0.984 | 0.016 | 1.635 | 88 | 0.9 | 0.01 |
| 1.169 | 1.222 | 0.982 | 0.018 | 1.793 | 89 | 0.9 | 0.01 |
| 1.222 | 1.278 | 0.980 | 0.020 | 1.977 | 90 | 0.9 | 0.01 |
| 1.278 | 1.338 | 0.978 | 0.022 | 2.196 | 91 | 0.9 | 0.01 |
| 1.338 | 1.404 | 0.975 | 0.025 | 2.459 | 92 | 0.9 | 0.01 |
| 1.404 | 1.476 | 0.972 | 0.028 | 2.783 | 93 | 0.9 | 0.01 |
| 1.476 | 1.558 | 0.968 | 0.032 | 3.193 | 94 | 0.9 | 0.01 |
| 1.558 | 1.651 | 0.963 | 0.037 | 3.728 | 95 | 0.9 | 0.01 |
| 1.651 | 1.761 | 0.955 | 0.045 | 4.459 | 96 | 0.9 | 0.01 |
| 1.761 | 1.898 | 0.945 | 0.055 | 5.529 | 97 | 0.9 | 0.01 |
| 1.898 | 2.082 | 0.927 | 0.073 | 7.271 | 98 | 0.9 | 0.01 |
| 2.082 | 2.380 | 0.892 | 0.108 | 10.768 | 99 | 0.9 | 0.01 |
| 2.380 | Inf | 0.734 | 0.266 | 26.623 | 100 | 0.9 | 0.01 |
| -Inf | -2.324 | 0.939 | 0.061 | 0.406 | 1 | 0.6 | 0.15 |
| -2.324 | -2.052 | 0.929 | 0.071 | 0.476 | 2 | 0.6 | 0.15 |
| -2.052 | -1.879 | 0.923 | 0.077 | 0.512 | 3 | 0.6 | 0.15 |
| -1.879 | -1.749 | 0.919 | 0.081 | 0.538 | 4 | 0.6 | 0.15 |
| -1.749 | -1.644 | 0.916 | 0.084 | 0.559 | 5 | 0.6 | 0.15 |
| -1.644 | -1.554 | 0.913 | 0.087 | 0.577 | 6 | 0.6 | 0.15 |
| -1.554 | -1.475 | 0.911 | 0.089 | 0.593 | 7 | 0.6 | 0.15 |
| -1.475 | -1.404 | 0.909 | 0.091 | 0.608 | 8 | 0.6 | 0.15 |
| -1.404 | -1.340 | 0.907 | 0.093 | 0.621 | 9 | 0.6 | 0.15 |
| -1.340 | -1.281 | 0.905 | 0.095 | 0.634 | 10 | 0.6 | 0.15 |
| -1.281 | -1.226 | 0.903 | 0.097 | 0.646 | 11 | 0.6 | 0.15 |
| -1.226 | -1.175 | 0.901 | 0.099 | 0.657 | 12 | 0.6 | 0.15 |
| -1.175 | -1.126 | 0.900 | 0.100 | 0.668 | 13 | 0.6 | 0.15 |
| -1.126 | -1.080 | 0.898 | 0.102 | 0.678 | 14 | 0.6 | 0.15 |
| -1.080 | -1.036 | 0.897 | 0.103 | 0.688 | 15 | 0.6 | 0.15 |
| -1.036 | -0.994 | 0.895 | 0.105 | 0.698 | 16 | 0.6 | 0.15 |
| -0.994 | -0.954 | 0.894 | 0.106 | 0.707 | 17 | 0.6 | 0.15 |
| -0.954 | -0.915 | 0.893 | 0.107 | 0.716 | 18 | 0.6 | 0.15 |
| -0.915 | -0.878 | 0.891 | 0.109 | 0.725 | 19 | 0.6 | 0.15 |
| -0.878 | -0.842 | 0.890 | 0.110 | 0.734 | 20 | 0.6 | 0.15 |
| -0.842 | -0.807 | 0.889 | 0.111 | 0.742 | 21 | 0.6 | 0.15 |
| -0.807 | -0.772 | 0.887 | 0.113 | 0.751 | 22 | 0.6 | 0.15 |
| -0.772 | -0.739 | 0.886 | 0.114 | 0.759 | 23 | 0.6 | 0.15 |
| -0.739 | -0.707 | 0.885 | 0.115 | 0.767 | 24 | 0.6 | 0.15 |
| -0.707 | -0.675 | 0.884 | 0.116 | 0.775 | 25 | 0.6 | 0.15 |
| -0.675 | -0.644 | 0.883 | 0.117 | 0.782 | 26 | 0.6 | 0.15 |
| -0.644 | -0.613 | 0.881 | 0.119 | 0.790 | 27 | 0.6 | 0.15 |
| -0.613 | -0.583 | 0.880 | 0.120 | 0.798 | 28 | 0.6 | 0.15 |
| -0.583 | -0.554 | 0.879 | 0.121 | 0.805 | 29 | 0.6 | 0.15 |
| -0.554 | -0.525 | 0.878 | 0.122 | 0.813 | 30 | 0.6 | 0.15 |
| -0.525 | -0.496 | 0.877 | 0.123 | 0.820 | 31 | 0.6 | 0.15 |
| -0.496 | -0.468 | 0.876 | 0.124 | 0.828 | 32 | 0.6 | 0.15 |
| -0.468 | -0.440 | 0.875 | 0.125 | 0.835 | 33 | 0.6 | 0.15 |
| -0.440 | -0.413 | 0.874 | 0.126 | 0.842 | 34 | 0.6 | 0.15 |
| -0.413 | -0.386 | 0.873 | 0.127 | 0.850 | 35 | 0.6 | 0.15 |
| -0.386 | -0.359 | 0.871 | 0.129 | 0.857 | 36 | 0.6 | 0.15 |
| -0.359 | -0.332 | 0.870 | 0.130 | 0.864 | 37 | 0.6 | 0.15 |
| -0.332 | -0.306 | 0.869 | 0.131 | 0.871 | 38 | 0.6 | 0.15 |
| -0.306 | -0.280 | 0.868 | 0.132 | 0.879 | 39 | 0.6 | 0.15 |
| -0.280 | -0.254 | 0.867 | 0.133 | 0.886 | 40 | 0.6 | 0.15 |
| -0.254 | -0.228 | 0.866 | 0.134 | 0.893 | 41 | 0.6 | 0.15 |
| -0.228 | -0.203 | 0.865 | 0.135 | 0.900 | 42 | 0.6 | 0.15 |
| -0.203 | -0.177 | 0.864 | 0.136 | 0.907 | 43 | 0.6 | 0.15 |
| -0.177 | -0.152 | 0.863 | 0.137 | 0.915 | 44 | 0.6 | 0.15 |
| -0.152 | -0.126 | 0.862 | 0.138 | 0.922 | 45 | 0.6 | 0.15 |
| -0.126 | -0.101 | 0.861 | 0.139 | 0.929 | 46 | 0.6 | 0.15 |
| -0.101 | -0.076 | 0.860 | 0.140 | 0.936 | 47 | 0.6 | 0.15 |
| -0.076 | -0.051 | 0.858 | 0.142 | 0.944 | 48 | 0.6 | 0.15 |
| -0.051 | -0.026 | 0.857 | 0.143 | 0.951 | 49 | 0.6 | 0.15 |
| -0.026 | -0.001 | 0.856 | 0.144 | 0.958 | 50 | 0.6 | 0.15 |
| -0.001 | 0.024 | 0.855 | 0.145 | 0.966 | 51 | 0.6 | 0.15 |
| 0.024 | 0.049 | 0.854 | 0.146 | 0.973 | 52 | 0.6 | 0.15 |
| 0.049 | 0.075 | 0.853 | 0.147 | 0.981 | 53 | 0.6 | 0.15 |
| 0.075 | 0.100 | 0.852 | 0.148 | 0.989 | 54 | 0.6 | 0.15 |
| 0.100 | 0.125 | 0.851 | 0.149 | 0.996 | 55 | 0.6 | 0.15 |
| 0.125 | 0.150 | 0.849 | 0.151 | 1.004 | 56 | 0.6 | 0.15 |
| 0.150 | 0.176 | 0.848 | 0.152 | 1.012 | 57 | 0.6 | 0.15 |
| 0.176 | 0.201 | 0.847 | 0.153 | 1.020 | 58 | 0.6 | 0.15 |
| 0.201 | 0.227 | 0.846 | 0.154 | 1.028 | 59 | 0.6 | 0.15 |
| 0.227 | 0.253 | 0.845 | 0.155 | 1.036 | 60 | 0.6 | 0.15 |
| 0.253 | 0.279 | 0.843 | 0.157 | 1.044 | 61 | 0.6 | 0.15 |
| 0.279 | 0.305 | 0.842 | 0.158 | 1.052 | 62 | 0.6 | 0.15 |
| 0.305 | 0.331 | 0.841 | 0.159 | 1.061 | 63 | 0.6 | 0.15 |
| 0.331 | 0.358 | 0.840 | 0.160 | 1.069 | 64 | 0.6 | 0.15 |
| 0.358 | 0.385 | 0.838 | 0.162 | 1.078 | 65 | 0.6 | 0.15 |
| 0.385 | 0.412 | 0.837 | 0.163 | 1.087 | 66 | 0.6 | 0.15 |
| 0.412 | 0.439 | 0.836 | 0.164 | 1.096 | 67 | 0.6 | 0.15 |
| 0.439 | 0.467 | 0.834 | 0.166 | 1.105 | 68 | 0.6 | 0.15 |
| 0.467 | 0.495 | 0.833 | 0.167 | 1.115 | 69 | 0.6 | 0.15 |
| 0.495 | 0.524 | 0.831 | 0.169 | 1.124 | 70 | 0.6 | 0.15 |
| 0.524 | 0.553 | 0.830 | 0.170 | 1.134 | 71 | 0.6 | 0.15 |
| 0.553 | 0.582 | 0.828 | 0.172 | 1.144 | 72 | 0.6 | 0.15 |
| 0.582 | 0.612 | 0.827 | 0.173 | 1.154 | 73 | 0.6 | 0.15 |
| 0.612 | 0.643 | 0.825 | 0.175 | 1.165 | 74 | 0.6 | 0.15 |
| 0.643 | 0.674 | 0.824 | 0.176 | 1.175 | 75 | 0.6 | 0.15 |
| 0.674 | 0.706 | 0.822 | 0.178 | 1.186 | 76 | 0.6 | 0.15 |
| 0.706 | 0.739 | 0.820 | 0.180 | 1.198 | 77 | 0.6 | 0.15 |
| 0.739 | 0.772 | 0.819 | 0.181 | 1.209 | 78 | 0.6 | 0.15 |
| 0.772 | 0.806 | 0.817 | 0.183 | 1.222 | 79 | 0.6 | 0.15 |
| 0.806 | 0.841 | 0.815 | 0.185 | 1.234 | 80 | 0.6 | 0.15 |
| 0.841 | 0.878 | 0.813 | 0.187 | 1.247 | 81 | 0.6 | 0.15 |
| 0.878 | 0.915 | 0.811 | 0.189 | 1.261 | 82 | 0.6 | 0.15 |
| 0.915 | 0.954 | 0.809 | 0.191 | 1.275 | 83 | 0.6 | 0.15 |
| 0.954 | 0.994 | 0.807 | 0.193 | 1.290 | 84 | 0.6 | 0.15 |
| 0.994 | 1.036 | 0.804 | 0.196 | 1.305 | 85 | 0.6 | 0.15 |
| 1.036 | 1.080 | 0.802 | 0.198 | 1.322 | 86 | 0.6 | 0.15 |
| 1.080 | 1.127 | 0.799 | 0.201 | 1.339 | 87 | 0.6 | 0.15 |
| 1.127 | 1.175 | 0.796 | 0.204 | 1.357 | 88 | 0.6 | 0.15 |
| 1.175 | 1.227 | 0.793 | 0.207 | 1.377 | 89 | 0.6 | 0.15 |
| 1.227 | 1.282 | 0.790 | 0.210 | 1.398 | 90 | 0.6 | 0.15 |
| 1.282 | 1.341 | 0.787 | 0.213 | 1.421 | 91 | 0.6 | 0.15 |
| 1.341 | 1.406 | 0.783 | 0.217 | 1.446 | 92 | 0.6 | 0.15 |
| 1.406 | 1.477 | 0.779 | 0.221 | 1.474 | 93 | 0.6 | 0.15 |
| 1.477 | 1.556 | 0.774 | 0.226 | 1.505 | 94 | 0.6 | 0.15 |
| 1.556 | 1.646 | 0.769 | 0.231 | 1.541 | 95 | 0.6 | 0.15 |
| 1.646 | 1.752 | 0.762 | 0.238 | 1.583 | 96 | 0.6 | 0.15 |
| 1.752 | 1.883 | 0.755 | 0.245 | 1.635 | 97 | 0.6 | 0.15 |
| 1.883 | 2.056 | 0.745 | 0.255 | 1.703 | 98 | 0.6 | 0.15 |
| 2.056 | 2.329 | 0.730 | 0.270 | 1.803 | 99 | 0.6 | 0.15 |
| 2.329 | Inf | 0.693 | 0.307 | 2.048 | 100 | 0.6 | 0.15 |
| -Inf | -2.304 | 0.981 | 0.019 | 0.129 | 1 | 0.7 | 0.15 |
| -2.304 | -2.037 | 0.973 | 0.027 | 0.181 | 2 | 0.7 | 0.15 |
| -2.037 | -1.867 | 0.968 | 0.032 | 0.211 | 3 | 0.7 | 0.15 |
| -1.867 | -1.739 | 0.965 | 0.035 | 0.235 | 4 | 0.7 | 0.15 |
| -1.739 | -1.635 | 0.962 | 0.038 | 0.256 | 5 | 0.7 | 0.15 |
| -1.635 | -1.546 | 0.959 | 0.041 | 0.275 | 6 | 0.7 | 0.15 |
| -1.546 | -1.469 | 0.956 | 0.044 | 0.292 | 7 | 0.7 | 0.15 |
| -1.469 | -1.399 | 0.954 | 0.046 | 0.308 | 8 | 0.7 | 0.15 |
| -1.399 | -1.336 | 0.951 | 0.049 | 0.324 | 9 | 0.7 | 0.15 |
| -1.336 | -1.277 | 0.949 | 0.051 | 0.338 | 10 | 0.7 | 0.15 |
| -1.277 | -1.223 | 0.947 | 0.053 | 0.352 | 11 | 0.7 | 0.15 |
| -1.223 | -1.172 | 0.945 | 0.055 | 0.366 | 12 | 0.7 | 0.15 |
| -1.172 | -1.124 | 0.943 | 0.057 | 0.379 | 13 | 0.7 | 0.15 |
| -1.124 | -1.079 | 0.941 | 0.059 | 0.392 | 14 | 0.7 | 0.15 |
| -1.079 | -1.035 | 0.939 | 0.061 | 0.405 | 15 | 0.7 | 0.15 |
| -1.035 | -0.994 | 0.937 | 0.063 | 0.418 | 16 | 0.7 | 0.15 |
| -0.994 | -0.954 | 0.935 | 0.065 | 0.430 | 17 | 0.7 | 0.15 |
| -0.954 | -0.915 | 0.934 | 0.066 | 0.442 | 18 | 0.7 | 0.15 |
| -0.915 | -0.878 | 0.932 | 0.068 | 0.454 | 19 | 0.7 | 0.15 |
| -0.878 | -0.842 | 0.930 | 0.070 | 0.466 | 20 | 0.7 | 0.15 |
| -0.842 | -0.808 | 0.928 | 0.072 | 0.478 | 21 | 0.7 | 0.15 |
| -0.808 | -0.774 | 0.926 | 0.074 | 0.490 | 22 | 0.7 | 0.15 |
| -0.774 | -0.741 | 0.925 | 0.075 | 0.502 | 23 | 0.7 | 0.15 |
| -0.741 | -0.708 | 0.923 | 0.077 | 0.514 | 24 | 0.7 | 0.15 |
| -0.708 | -0.677 | 0.921 | 0.079 | 0.526 | 25 | 0.7 | 0.15 |
| -0.677 | -0.646 | 0.919 | 0.081 | 0.537 | 26 | 0.7 | 0.15 |
| -0.646 | -0.616 | 0.918 | 0.082 | 0.549 | 27 | 0.7 | 0.15 |
| -0.616 | -0.586 | 0.916 | 0.084 | 0.561 | 28 | 0.7 | 0.15 |
| -0.586 | -0.557 | 0.914 | 0.086 | 0.572 | 29 | 0.7 | 0.15 |
| -0.557 | -0.528 | 0.912 | 0.088 | 0.584 | 30 | 0.7 | 0.15 |
| -0.528 | -0.500 | 0.911 | 0.089 | 0.596 | 31 | 0.7 | 0.15 |
| -0.500 | -0.472 | 0.909 | 0.091 | 0.608 | 32 | 0.7 | 0.15 |
| -0.472 | -0.444 | 0.907 | 0.093 | 0.620 | 33 | 0.7 | 0.15 |
| -0.444 | -0.417 | 0.905 | 0.095 | 0.632 | 34 | 0.7 | 0.15 |
| -0.417 | -0.390 | 0.903 | 0.097 | 0.644 | 35 | 0.7 | 0.15 |
| -0.390 | -0.363 | 0.902 | 0.098 | 0.656 | 36 | 0.7 | 0.15 |
| -0.363 | -0.337 | 0.900 | 0.100 | 0.668 | 37 | 0.7 | 0.15 |
| -0.337 | -0.310 | 0.898 | 0.102 | 0.680 | 38 | 0.7 | 0.15 |
| -0.310 | -0.284 | 0.896 | 0.104 | 0.692 | 39 | 0.7 | 0.15 |
| -0.284 | -0.258 | 0.894 | 0.106 | 0.705 | 40 | 0.7 | 0.15 |
| -0.258 | -0.233 | 0.892 | 0.108 | 0.717 | 41 | 0.7 | 0.15 |
| -0.233 | -0.207 | 0.890 | 0.110 | 0.730 | 42 | 0.7 | 0.15 |
| -0.207 | -0.182 | 0.889 | 0.111 | 0.743 | 43 | 0.7 | 0.15 |
| -0.182 | -0.156 | 0.887 | 0.113 | 0.756 | 44 | 0.7 | 0.15 |
| -0.156 | -0.131 | 0.885 | 0.115 | 0.769 | 45 | 0.7 | 0.15 |
| -0.131 | -0.106 | 0.883 | 0.117 | 0.782 | 46 | 0.7 | 0.15 |
| -0.106 | -0.081 | 0.881 | 0.119 | 0.796 | 47 | 0.7 | 0.15 |
| -0.081 | -0.056 | 0.879 | 0.121 | 0.809 | 48 | 0.7 | 0.15 |
| -0.056 | -0.031 | 0.877 | 0.123 | 0.823 | 49 | 0.7 | 0.15 |
| -0.031 | -0.006 | 0.874 | 0.126 | 0.837 | 50 | 0.7 | 0.15 |
| -0.006 | 0.019 | 0.872 | 0.128 | 0.851 | 51 | 0.7 | 0.15 |
| 0.019 | 0.044 | 0.870 | 0.130 | 0.865 | 52 | 0.7 | 0.15 |
| 0.044 | 0.070 | 0.868 | 0.132 | 0.880 | 53 | 0.7 | 0.15 |
| 0.070 | 0.095 | 0.866 | 0.134 | 0.895 | 54 | 0.7 | 0.15 |
| 0.095 | 0.120 | 0.864 | 0.136 | 0.910 | 55 | 0.7 | 0.15 |
| 0.120 | 0.145 | 0.861 | 0.139 | 0.925 | 56 | 0.7 | 0.15 |
| 0.145 | 0.171 | 0.859 | 0.141 | 0.941 | 57 | 0.7 | 0.15 |
| 0.171 | 0.196 | 0.856 | 0.144 | 0.957 | 58 | 0.7 | 0.15 |
| 0.196 | 0.222 | 0.854 | 0.146 | 0.973 | 59 | 0.7 | 0.15 |
| 0.222 | 0.248 | 0.852 | 0.148 | 0.990 | 60 | 0.7 | 0.15 |
| 0.248 | 0.274 | 0.849 | 0.151 | 1.006 | 61 | 0.7 | 0.15 |
| 0.274 | 0.300 | 0.846 | 0.154 | 1.024 | 62 | 0.7 | 0.15 |
| 0.300 | 0.326 | 0.844 | 0.156 | 1.041 | 63 | 0.7 | 0.15 |
| 0.326 | 0.353 | 0.841 | 0.159 | 1.059 | 64 | 0.7 | 0.15 |
| 0.353 | 0.380 | 0.838 | 0.162 | 1.078 | 65 | 0.7 | 0.15 |
| 0.380 | 0.407 | 0.835 | 0.165 | 1.097 | 66 | 0.7 | 0.15 |
| 0.407 | 0.435 | 0.833 | 0.167 | 1.116 | 67 | 0.7 | 0.15 |
| 0.435 | 0.463 | 0.830 | 0.170 | 1.136 | 68 | 0.7 | 0.15 |
| 0.463 | 0.491 | 0.827 | 0.173 | 1.156 | 69 | 0.7 | 0.15 |
| 0.491 | 0.520 | 0.823 | 0.177 | 1.178 | 70 | 0.7 | 0.15 |
| 0.520 | 0.549 | 0.820 | 0.180 | 1.199 | 71 | 0.7 | 0.15 |
| 0.549 | 0.579 | 0.817 | 0.183 | 1.222 | 72 | 0.7 | 0.15 |
| 0.579 | 0.609 | 0.813 | 0.187 | 1.245 | 73 | 0.7 | 0.15 |
| 0.609 | 0.639 | 0.810 | 0.190 | 1.268 | 74 | 0.7 | 0.15 |
| 0.639 | 0.671 | 0.806 | 0.194 | 1.293 | 75 | 0.7 | 0.15 |
| 0.671 | 0.703 | 0.802 | 0.198 | 1.319 | 76 | 0.7 | 0.15 |
| 0.703 | 0.736 | 0.798 | 0.202 | 1.345 | 77 | 0.7 | 0.15 |
| 0.736 | 0.769 | 0.794 | 0.206 | 1.373 | 78 | 0.7 | 0.15 |
| 0.769 | 0.804 | 0.790 | 0.210 | 1.401 | 79 | 0.7 | 0.15 |
| 0.804 | 0.839 | 0.785 | 0.215 | 1.431 | 80 | 0.7 | 0.15 |
| 0.839 | 0.876 | 0.781 | 0.219 | 1.463 | 81 | 0.7 | 0.15 |
| 0.876 | 0.914 | 0.776 | 0.224 | 1.496 | 82 | 0.7 | 0.15 |
| 0.914 | 0.953 | 0.770 | 0.230 | 1.530 | 83 | 0.7 | 0.15 |
| 0.953 | 0.994 | 0.765 | 0.235 | 1.567 | 84 | 0.7 | 0.15 |
| 0.994 | 1.036 | 0.759 | 0.241 | 1.605 | 85 | 0.7 | 0.15 |
| 1.036 | 1.081 | 0.753 | 0.247 | 1.646 | 86 | 0.7 | 0.15 |
| 1.081 | 1.127 | 0.747 | 0.253 | 1.690 | 87 | 0.7 | 0.15 |
| 1.127 | 1.177 | 0.740 | 0.260 | 1.737 | 88 | 0.7 | 0.15 |
| 1.177 | 1.229 | 0.732 | 0.268 | 1.787 | 89 | 0.7 | 0.15 |
| 1.229 | 1.285 | 0.724 | 0.276 | 1.842 | 90 | 0.7 | 0.15 |
| 1.285 | 1.345 | 0.715 | 0.285 | 1.902 | 91 | 0.7 | 0.15 |
| 1.345 | 1.410 | 0.705 | 0.295 | 1.968 | 92 | 0.7 | 0.15 |
| 1.410 | 1.482 | 0.694 | 0.306 | 2.041 | 93 | 0.7 | 0.15 |
| 1.482 | 1.563 | 0.681 | 0.319 | 2.125 | 94 | 0.7 | 0.15 |
| 1.563 | 1.655 | 0.667 | 0.333 | 2.221 | 95 | 0.7 | 0.15 |
| 1.655 | 1.763 | 0.650 | 0.350 | 2.336 | 96 | 0.7 | 0.15 |
| 1.763 | 1.896 | 0.628 | 0.372 | 2.477 | 97 | 0.7 | 0.15 |
| 1.896 | 2.073 | 0.601 | 0.399 | 2.662 | 98 | 0.7 | 0.15 |
| 2.073 | 2.352 | 0.560 | 0.440 | 2.935 | 99 | 0.7 | 0.15 |
| 2.352 | Inf | 0.465 | 0.535 | 3.568 | 100 | 0.7 | 0.15 |
| -Inf | -2.250 | 0.996 | 0.004 | 0.027 | 1 | 0.8 | 0.15 |
| -2.250 | -1.994 | 0.993 | 0.007 | 0.047 | 2 | 0.8 | 0.15 |
| -1.994 | -1.831 | 0.991 | 0.009 | 0.060 | 3 | 0.8 | 0.15 |
| -1.831 | -1.708 | 0.989 | 0.011 | 0.072 | 4 | 0.8 | 0.15 |
| -1.708 | -1.608 | 0.988 | 0.012 | 0.083 | 5 | 0.8 | 0.15 |
| -1.608 | -1.523 | 0.986 | 0.014 | 0.093 | 6 | 0.8 | 0.15 |
| -1.523 | -1.448 | 0.984 | 0.016 | 0.103 | 7 | 0.8 | 0.15 |
| -1.448 | -1.381 | 0.983 | 0.017 | 0.113 | 8 | 0.8 | 0.15 |
| -1.381 | -1.320 | 0.982 | 0.018 | 0.123 | 9 | 0.8 | 0.15 |
| -1.320 | -1.264 | 0.980 | 0.020 | 0.132 | 10 | 0.8 | 0.15 |
| -1.264 | -1.211 | 0.979 | 0.021 | 0.142 | 11 | 0.8 | 0.15 |
| -1.211 | -1.162 | 0.977 | 0.023 | 0.151 | 12 | 0.8 | 0.15 |
| -1.162 | -1.116 | 0.976 | 0.024 | 0.160 | 13 | 0.8 | 0.15 |
| -1.116 | -1.072 | 0.975 | 0.025 | 0.170 | 14 | 0.8 | 0.15 |
| -1.072 | -1.030 | 0.973 | 0.027 | 0.179 | 15 | 0.8 | 0.15 |
| -1.030 | -0.989 | 0.972 | 0.028 | 0.189 | 16 | 0.8 | 0.15 |
| -0.989 | -0.951 | 0.970 | 0.030 | 0.198 | 17 | 0.8 | 0.15 |
| -0.951 | -0.914 | 0.969 | 0.031 | 0.208 | 18 | 0.8 | 0.15 |
| -0.914 | -0.878 | 0.967 | 0.033 | 0.218 | 19 | 0.8 | 0.15 |
| -0.878 | -0.843 | 0.966 | 0.034 | 0.228 | 20 | 0.8 | 0.15 |
| -0.843 | -0.809 | 0.964 | 0.036 | 0.238 | 21 | 0.8 | 0.15 |
| -0.809 | -0.776 | 0.963 | 0.037 | 0.248 | 22 | 0.8 | 0.15 |
| -0.776 | -0.744 | 0.961 | 0.039 | 0.258 | 23 | 0.8 | 0.15 |
| -0.744 | -0.712 | 0.960 | 0.040 | 0.268 | 24 | 0.8 | 0.15 |
| -0.712 | -0.681 | 0.958 | 0.042 | 0.279 | 25 | 0.8 | 0.15 |
| -0.681 | -0.651 | 0.957 | 0.043 | 0.290 | 26 | 0.8 | 0.15 |
| -0.651 | -0.622 | 0.955 | 0.045 | 0.300 | 27 | 0.8 | 0.15 |
| -0.622 | -0.593 | 0.953 | 0.047 | 0.311 | 28 | 0.8 | 0.15 |
| -0.593 | -0.564 | 0.952 | 0.048 | 0.323 | 29 | 0.8 | 0.15 |
| -0.564 | -0.536 | 0.950 | 0.050 | 0.334 | 30 | 0.8 | 0.15 |
| -0.536 | -0.508 | 0.948 | 0.052 | 0.346 | 31 | 0.8 | 0.15 |
| -0.508 | -0.481 | 0.946 | 0.054 | 0.358 | 32 | 0.8 | 0.15 |
| -0.481 | -0.454 | 0.945 | 0.055 | 0.370 | 33 | 0.8 | 0.15 |
| -0.454 | -0.427 | 0.943 | 0.057 | 0.382 | 34 | 0.8 | 0.15 |
| -0.427 | -0.401 | 0.941 | 0.059 | 0.395 | 35 | 0.8 | 0.15 |
| -0.401 | -0.374 | 0.939 | 0.061 | 0.408 | 36 | 0.8 | 0.15 |
| -0.374 | -0.348 | 0.937 | 0.063 | 0.421 | 37 | 0.8 | 0.15 |
| -0.348 | -0.322 | 0.935 | 0.065 | 0.434 | 38 | 0.8 | 0.15 |
| -0.322 | -0.297 | 0.933 | 0.067 | 0.448 | 39 | 0.8 | 0.15 |
| -0.297 | -0.271 | 0.931 | 0.069 | 0.462 | 40 | 0.8 | 0.15 |
| -0.271 | -0.246 | 0.929 | 0.071 | 0.476 | 41 | 0.8 | 0.15 |
| -0.246 | -0.221 | 0.926 | 0.074 | 0.491 | 42 | 0.8 | 0.15 |
| -0.221 | -0.196 | 0.924 | 0.076 | 0.506 | 43 | 0.8 | 0.15 |
| -0.196 | -0.171 | 0.922 | 0.078 | 0.521 | 44 | 0.8 | 0.15 |
| -0.171 | -0.146 | 0.919 | 0.081 | 0.537 | 45 | 0.8 | 0.15 |
| -0.146 | -0.121 | 0.917 | 0.083 | 0.553 | 46 | 0.8 | 0.15 |
| -0.121 | -0.096 | 0.915 | 0.085 | 0.570 | 47 | 0.8 | 0.15 |
| -0.096 | -0.071 | 0.912 | 0.088 | 0.587 | 48 | 0.8 | 0.15 |
| -0.071 | -0.046 | 0.909 | 0.091 | 0.604 | 49 | 0.8 | 0.15 |
| -0.046 | -0.021 | 0.907 | 0.093 | 0.622 | 50 | 0.8 | 0.15 |
| -0.021 | 0.003 | 0.904 | 0.096 | 0.640 | 51 | 0.8 | 0.15 |
| 0.003 | 0.028 | 0.901 | 0.099 | 0.659 | 52 | 0.8 | 0.15 |
| 0.028 | 0.053 | 0.898 | 0.102 | 0.679 | 53 | 0.8 | 0.15 |
| 0.053 | 0.078 | 0.895 | 0.105 | 0.699 | 54 | 0.8 | 0.15 |
| 0.078 | 0.104 | 0.892 | 0.108 | 0.719 | 55 | 0.8 | 0.15 |
| 0.104 | 0.129 | 0.889 | 0.111 | 0.740 | 56 | 0.8 | 0.15 |
| 0.129 | 0.154 | 0.886 | 0.114 | 0.762 | 57 | 0.8 | 0.15 |
| 0.154 | 0.180 | 0.882 | 0.118 | 0.785 | 58 | 0.8 | 0.15 |
| 0.180 | 0.206 | 0.879 | 0.121 | 0.808 | 59 | 0.8 | 0.15 |
| 0.206 | 0.231 | 0.875 | 0.125 | 0.832 | 60 | 0.8 | 0.15 |
| 0.231 | 0.258 | 0.871 | 0.129 | 0.857 | 61 | 0.8 | 0.15 |
| 0.258 | 0.284 | 0.868 | 0.132 | 0.882 | 62 | 0.8 | 0.15 |
| 0.284 | 0.311 | 0.864 | 0.136 | 0.909 | 63 | 0.8 | 0.15 |
| 0.311 | 0.337 | 0.860 | 0.140 | 0.936 | 64 | 0.8 | 0.15 |
| 0.337 | 0.365 | 0.855 | 0.145 | 0.965 | 65 | 0.8 | 0.15 |
| 0.365 | 0.392 | 0.851 | 0.149 | 0.994 | 66 | 0.8 | 0.15 |
| 0.392 | 0.420 | 0.846 | 0.154 | 1.025 | 67 | 0.8 | 0.15 |
| 0.420 | 0.448 | 0.841 | 0.159 | 1.057 | 68 | 0.8 | 0.15 |
| 0.448 | 0.477 | 0.837 | 0.163 | 1.090 | 69 | 0.8 | 0.15 |
| 0.477 | 0.506 | 0.831 | 0.169 | 1.124 | 70 | 0.8 | 0.15 |
| 0.506 | 0.535 | 0.826 | 0.174 | 1.160 | 71 | 0.8 | 0.15 |
| 0.535 | 0.565 | 0.820 | 0.180 | 1.198 | 72 | 0.8 | 0.15 |
| 0.565 | 0.596 | 0.814 | 0.186 | 1.237 | 73 | 0.8 | 0.15 |
| 0.596 | 0.627 | 0.808 | 0.192 | 1.278 | 74 | 0.8 | 0.15 |
| 0.627 | 0.659 | 0.802 | 0.198 | 1.320 | 75 | 0.8 | 0.15 |
| 0.659 | 0.692 | 0.795 | 0.205 | 1.365 | 76 | 0.8 | 0.15 |
| 0.692 | 0.726 | 0.788 | 0.212 | 1.412 | 77 | 0.8 | 0.15 |
| 0.726 | 0.760 | 0.781 | 0.219 | 1.462 | 78 | 0.8 | 0.15 |
| 0.760 | 0.796 | 0.773 | 0.227 | 1.514 | 79 | 0.8 | 0.15 |
| 0.796 | 0.832 | 0.765 | 0.235 | 1.569 | 80 | 0.8 | 0.15 |
| 0.832 | 0.870 | 0.756 | 0.244 | 1.628 | 81 | 0.8 | 0.15 |
| 0.870 | 0.909 | 0.747 | 0.253 | 1.689 | 82 | 0.8 | 0.15 |
| 0.909 | 0.949 | 0.737 | 0.263 | 1.755 | 83 | 0.8 | 0.15 |
| 0.949 | 0.991 | 0.726 | 0.274 | 1.825 | 84 | 0.8 | 0.15 |
| 0.991 | 1.035 | 0.715 | 0.285 | 1.900 | 85 | 0.8 | 0.15 |
| 1.035 | 1.081 | 0.703 | 0.297 | 1.980 | 86 | 0.8 | 0.15 |
| 1.081 | 1.130 | 0.690 | 0.310 | 2.066 | 87 | 0.8 | 0.15 |
| 1.130 | 1.181 | 0.676 | 0.324 | 2.159 | 88 | 0.8 | 0.15 |
| 1.181 | 1.236 | 0.661 | 0.339 | 2.260 | 89 | 0.8 | 0.15 |
| 1.236 | 1.294 | 0.644 | 0.356 | 2.370 | 90 | 0.8 | 0.15 |
| 1.294 | 1.357 | 0.626 | 0.374 | 2.492 | 91 | 0.8 | 0.15 |
| 1.357 | 1.426 | 0.606 | 0.394 | 2.626 | 92 | 0.8 | 0.15 |
| 1.426 | 1.502 | 0.584 | 0.416 | 2.775 | 93 | 0.8 | 0.15 |
| 1.502 | 1.587 | 0.558 | 0.442 | 2.944 | 94 | 0.8 | 0.15 |
| 1.587 | 1.684 | 0.529 | 0.471 | 3.138 | 95 | 0.8 | 0.15 |
| 1.684 | 1.798 | 0.495 | 0.505 | 3.364 | 96 | 0.8 | 0.15 |
| 1.798 | 1.939 | 0.455 | 0.545 | 3.636 | 97 | 0.8 | 0.15 |
| 1.939 | 2.126 | 0.404 | 0.596 | 3.976 | 98 | 0.8 | 0.15 |
| 2.126 | 2.422 | 0.334 | 0.666 | 4.439 | 99 | 0.8 | 0.15 |
| 2.422 | Inf | 0.211 | 0.789 | 5.263 | 100 | 0.8 | 0.15 |
| -Inf | -2.130 | 1.000 | 0.000 | 0.002 | 1 | 0.9 | 0.15 |
| -2.130 | -1.896 | 0.999 | 0.001 | 0.005 | 2 | 0.9 | 0.15 |
| -1.896 | -1.746 | 0.999 | 0.001 | 0.007 | 3 | 0.9 | 0.15 |
| -1.746 | -1.634 | 0.999 | 0.001 | 0.010 | 4 | 0.9 | 0.15 |
| -1.634 | -1.542 | 0.998 | 0.002 | 0.012 | 5 | 0.9 | 0.15 |
| -1.542 | -1.464 | 0.998 | 0.002 | 0.015 | 6 | 0.9 | 0.15 |
| -1.464 | -1.395 | 0.997 | 0.003 | 0.017 | 7 | 0.9 | 0.15 |
| -1.395 | -1.334 | 0.997 | 0.003 | 0.020 | 8 | 0.9 | 0.15 |
| -1.334 | -1.277 | 0.997 | 0.003 | 0.022 | 9 | 0.9 | 0.15 |
| -1.277 | -1.226 | 0.996 | 0.004 | 0.025 | 10 | 0.9 | 0.15 |
| -1.226 | -1.177 | 0.996 | 0.004 | 0.028 | 11 | 0.9 | 0.15 |
| -1.177 | -1.132 | 0.995 | 0.005 | 0.031 | 12 | 0.9 | 0.15 |
| -1.132 | -1.089 | 0.995 | 0.005 | 0.034 | 13 | 0.9 | 0.15 |
| -1.089 | -1.049 | 0.994 | 0.006 | 0.037 | 14 | 0.9 | 0.15 |
| -1.049 | -1.010 | 0.994 | 0.006 | 0.040 | 15 | 0.9 | 0.15 |
| -1.010 | -0.973 | 0.993 | 0.007 | 0.044 | 16 | 0.9 | 0.15 |
| -0.973 | -0.937 | 0.993 | 0.007 | 0.047 | 17 | 0.9 | 0.15 |
| -0.937 | -0.902 | 0.992 | 0.008 | 0.051 | 18 | 0.9 | 0.15 |
| -0.902 | -0.869 | 0.992 | 0.008 | 0.055 | 19 | 0.9 | 0.15 |
| -0.869 | -0.837 | 0.991 | 0.009 | 0.059 | 20 | 0.9 | 0.15 |
| -0.837 | -0.805 | 0.991 | 0.009 | 0.063 | 21 | 0.9 | 0.15 |
| -0.805 | -0.775 | 0.990 | 0.010 | 0.067 | 22 | 0.9 | 0.15 |
| -0.775 | -0.745 | 0.989 | 0.011 | 0.072 | 23 | 0.9 | 0.15 |
| -0.745 | -0.715 | 0.989 | 0.011 | 0.076 | 24 | 0.9 | 0.15 |
| -0.715 | -0.687 | 0.988 | 0.012 | 0.081 | 25 | 0.9 | 0.15 |
| -0.687 | -0.659 | 0.987 | 0.013 | 0.086 | 26 | 0.9 | 0.15 |
| -0.659 | -0.631 | 0.986 | 0.014 | 0.091 | 27 | 0.9 | 0.15 |
| -0.631 | -0.604 | 0.985 | 0.015 | 0.097 | 28 | 0.9 | 0.15 |
| -0.604 | -0.577 | 0.985 | 0.015 | 0.103 | 29 | 0.9 | 0.15 |
| -0.577 | -0.551 | 0.984 | 0.016 | 0.109 | 30 | 0.9 | 0.15 |
| -0.551 | -0.525 | 0.983 | 0.017 | 0.115 | 31 | 0.9 | 0.15 |
| -0.525 | -0.499 | 0.982 | 0.018 | 0.121 | 32 | 0.9 | 0.15 |
| -0.499 | -0.473 | 0.981 | 0.019 | 0.128 | 33 | 0.9 | 0.15 |
| -0.473 | -0.448 | 0.980 | 0.020 | 0.135 | 34 | 0.9 | 0.15 |
| -0.448 | -0.423 | 0.979 | 0.021 | 0.142 | 35 | 0.9 | 0.15 |
| -0.423 | -0.398 | 0.977 | 0.023 | 0.150 | 36 | 0.9 | 0.15 |
| -0.398 | -0.374 | 0.976 | 0.024 | 0.158 | 37 | 0.9 | 0.15 |
| -0.374 | -0.349 | 0.975 | 0.025 | 0.166 | 38 | 0.9 | 0.15 |
| -0.349 | -0.325 | 0.974 | 0.026 | 0.175 | 39 | 0.9 | 0.15 |
| -0.325 | -0.301 | 0.972 | 0.028 | 0.184 | 40 | 0.9 | 0.15 |
| -0.301 | -0.277 | 0.971 | 0.029 | 0.194 | 41 | 0.9 | 0.15 |
| -0.277 | -0.253 | 0.969 | 0.031 | 0.204 | 42 | 0.9 | 0.15 |
| -0.253 | -0.229 | 0.968 | 0.032 | 0.214 | 43 | 0.9 | 0.15 |
| -0.229 | -0.205 | 0.966 | 0.034 | 0.225 | 44 | 0.9 | 0.15 |
| -0.205 | -0.181 | 0.964 | 0.036 | 0.237 | 45 | 0.9 | 0.15 |
| -0.181 | -0.157 | 0.963 | 0.037 | 0.249 | 46 | 0.9 | 0.15 |
| -0.157 | -0.133 | 0.961 | 0.039 | 0.262 | 47 | 0.9 | 0.15 |
| -0.133 | -0.109 | 0.959 | 0.041 | 0.275 | 48 | 0.9 | 0.15 |
| -0.109 | -0.085 | 0.957 | 0.043 | 0.289 | 49 | 0.9 | 0.15 |
| -0.085 | -0.061 | 0.954 | 0.046 | 0.304 | 50 | 0.9 | 0.15 |
| -0.061 | -0.037 | 0.952 | 0.048 | 0.319 | 51 | 0.9 | 0.15 |
| -0.037 | -0.013 | 0.950 | 0.050 | 0.335 | 52 | 0.9 | 0.15 |
| -0.013 | 0.011 | 0.947 | 0.053 | 0.352 | 53 | 0.9 | 0.15 |
| 0.011 | 0.036 | 0.944 | 0.056 | 0.370 | 54 | 0.9 | 0.15 |
| 0.036 | 0.060 | 0.942 | 0.058 | 0.389 | 55 | 0.9 | 0.15 |
| 0.060 | 0.085 | 0.939 | 0.061 | 0.409 | 56 | 0.9 | 0.15 |
| 0.085 | 0.110 | 0.935 | 0.065 | 0.430 | 57 | 0.9 | 0.15 |
| 0.110 | 0.135 | 0.932 | 0.068 | 0.453 | 58 | 0.9 | 0.15 |
| 0.135 | 0.161 | 0.929 | 0.071 | 0.476 | 59 | 0.9 | 0.15 |
| 0.161 | 0.187 | 0.925 | 0.075 | 0.501 | 60 | 0.9 | 0.15 |
| 0.187 | 0.212 | 0.921 | 0.079 | 0.528 | 61 | 0.9 | 0.15 |
| 0.212 | 0.239 | 0.917 | 0.083 | 0.556 | 62 | 0.9 | 0.15 |
| 0.239 | 0.265 | 0.912 | 0.088 | 0.586 | 63 | 0.9 | 0.15 |
| 0.265 | 0.292 | 0.907 | 0.093 | 0.617 | 64 | 0.9 | 0.15 |
| 0.292 | 0.320 | 0.902 | 0.098 | 0.651 | 65 | 0.9 | 0.15 |
| 0.320 | 0.347 | 0.897 | 0.103 | 0.687 | 66 | 0.9 | 0.15 |
| 0.347 | 0.376 | 0.891 | 0.109 | 0.725 | 67 | 0.9 | 0.15 |
| 0.376 | 0.404 | 0.885 | 0.115 | 0.766 | 68 | 0.9 | 0.15 |
| 0.404 | 0.434 | 0.879 | 0.121 | 0.809 | 69 | 0.9 | 0.15 |
| 0.434 | 0.464 | 0.872 | 0.128 | 0.856 | 70 | 0.9 | 0.15 |
| 0.464 | 0.494 | 0.864 | 0.136 | 0.905 | 71 | 0.9 | 0.15 |
| 0.494 | 0.525 | 0.856 | 0.144 | 0.959 | 72 | 0.9 | 0.15 |
| 0.525 | 0.557 | 0.848 | 0.152 | 1.016 | 73 | 0.9 | 0.15 |
| 0.557 | 0.590 | 0.838 | 0.162 | 1.078 | 74 | 0.9 | 0.15 |
| 0.590 | 0.624 | 0.828 | 0.172 | 1.144 | 75 | 0.9 | 0.15 |
| 0.624 | 0.658 | 0.818 | 0.182 | 1.215 | 76 | 0.9 | 0.15 |
| 0.658 | 0.694 | 0.806 | 0.194 | 1.293 | 77 | 0.9 | 0.15 |
| 0.694 | 0.731 | 0.794 | 0.206 | 1.376 | 78 | 0.9 | 0.15 |
| 0.731 | 0.769 | 0.780 | 0.220 | 1.466 | 79 | 0.9 | 0.15 |
| 0.769 | 0.808 | 0.765 | 0.235 | 1.564 | 80 | 0.9 | 0.15 |
| 0.808 | 0.849 | 0.749 | 0.251 | 1.670 | 81 | 0.9 | 0.15 |
| 0.849 | 0.892 | 0.732 | 0.268 | 1.786 | 82 | 0.9 | 0.15 |
| 0.892 | 0.936 | 0.713 | 0.287 | 1.912 | 83 | 0.9 | 0.15 |
| 0.936 | 0.983 | 0.693 | 0.307 | 2.049 | 84 | 0.9 | 0.15 |
| 0.983 | 1.032 | 0.670 | 0.330 | 2.198 | 85 | 0.9 | 0.15 |
| 1.032 | 1.084 | 0.646 | 0.354 | 2.361 | 86 | 0.9 | 0.15 |
| 1.084 | 1.138 | 0.619 | 0.381 | 2.538 | 87 | 0.9 | 0.15 |
| 1.138 | 1.197 | 0.590 | 0.410 | 2.732 | 88 | 0.9 | 0.15 |
| 1.197 | 1.259 | 0.558 | 0.442 | 2.944 | 89 | 0.9 | 0.15 |
| 1.259 | 1.325 | 0.524 | 0.476 | 3.174 | 90 | 0.9 | 0.15 |
| 1.325 | 1.398 | 0.486 | 0.514 | 3.424 | 91 | 0.9 | 0.15 |
| 1.398 | 1.477 | 0.446 | 0.554 | 3.694 | 92 | 0.9 | 0.15 |
| 1.477 | 1.564 | 0.402 | 0.598 | 3.984 | 93 | 0.9 | 0.15 |
| 1.564 | 1.661 | 0.356 | 0.644 | 4.295 | 94 | 0.9 | 0.15 |
| 1.661 | 1.771 | 0.306 | 0.694 | 4.624 | 95 | 0.9 | 0.15 |
| 1.771 | 1.900 | 0.255 | 0.745 | 4.968 | 96 | 0.9 | 0.15 |
| 1.900 | 2.057 | 0.201 | 0.799 | 5.324 | 97 | 0.9 | 0.15 |
| 2.057 | 2.260 | 0.147 | 0.853 | 5.687 | 98 | 0.9 | 0.15 |
| 2.260 | 2.570 | 0.092 | 0.908 | 6.050 | 99 | 0.9 | 0.15 |
| 2.570 | Inf | 0.037 | 0.963 | 6.423 | 100 | 0.9 | 0.15 |
| -Inf | -2.323 | 0.862 | 0.138 | 0.459 | 1 | 0.6 | 0.30 |
| -2.323 | -2.052 | 0.841 | 0.159 | 0.532 | 2 | 0.6 | 0.30 |
| -2.052 | -1.879 | 0.830 | 0.170 | 0.567 | 3 | 0.6 | 0.30 |
| -1.879 | -1.749 | 0.822 | 0.178 | 0.593 | 4 | 0.6 | 0.30 |
| -1.749 | -1.644 | 0.816 | 0.184 | 0.614 | 5 | 0.6 | 0.30 |
| -1.644 | -1.554 | 0.810 | 0.190 | 0.632 | 6 | 0.6 | 0.30 |
| -1.554 | -1.475 | 0.806 | 0.194 | 0.648 | 7 | 0.6 | 0.30 |
| -1.475 | -1.404 | 0.801 | 0.199 | 0.662 | 8 | 0.6 | 0.30 |
| -1.404 | -1.340 | 0.798 | 0.202 | 0.675 | 9 | 0.6 | 0.30 |
| -1.340 | -1.281 | 0.794 | 0.206 | 0.687 | 10 | 0.6 | 0.30 |
| -1.281 | -1.226 | 0.791 | 0.209 | 0.698 | 11 | 0.6 | 0.30 |
| -1.226 | -1.175 | 0.787 | 0.213 | 0.709 | 12 | 0.6 | 0.30 |
| -1.175 | -1.126 | 0.784 | 0.216 | 0.719 | 13 | 0.6 | 0.30 |
| -1.126 | -1.080 | 0.781 | 0.219 | 0.729 | 14 | 0.6 | 0.30 |
| -1.080 | -1.036 | 0.779 | 0.221 | 0.738 | 15 | 0.6 | 0.30 |
| -1.036 | -0.995 | 0.776 | 0.224 | 0.747 | 16 | 0.6 | 0.30 |
| -0.995 | -0.954 | 0.773 | 0.227 | 0.756 | 17 | 0.6 | 0.30 |
| -0.954 | -0.916 | 0.771 | 0.229 | 0.764 | 18 | 0.6 | 0.30 |
| -0.916 | -0.878 | 0.768 | 0.232 | 0.772 | 19 | 0.6 | 0.30 |
| -0.878 | -0.842 | 0.766 | 0.234 | 0.780 | 20 | 0.6 | 0.30 |
| -0.842 | -0.807 | 0.764 | 0.236 | 0.788 | 21 | 0.6 | 0.30 |
| -0.807 | -0.773 | 0.761 | 0.239 | 0.796 | 22 | 0.6 | 0.30 |
| -0.773 | -0.739 | 0.759 | 0.241 | 0.803 | 23 | 0.6 | 0.30 |
| -0.739 | -0.707 | 0.757 | 0.243 | 0.811 | 24 | 0.6 | 0.30 |
| -0.707 | -0.675 | 0.755 | 0.245 | 0.818 | 25 | 0.6 | 0.30 |
| -0.675 | -0.644 | 0.753 | 0.247 | 0.825 | 26 | 0.6 | 0.30 |
| -0.644 | -0.613 | 0.750 | 0.250 | 0.832 | 27 | 0.6 | 0.30 |
| -0.613 | -0.583 | 0.748 | 0.252 | 0.839 | 28 | 0.6 | 0.30 |
| -0.583 | -0.554 | 0.746 | 0.254 | 0.845 | 29 | 0.6 | 0.30 |
| -0.554 | -0.525 | 0.744 | 0.256 | 0.852 | 30 | 0.6 | 0.30 |
| -0.525 | -0.496 | 0.742 | 0.258 | 0.859 | 31 | 0.6 | 0.30 |
| -0.496 | -0.468 | 0.740 | 0.260 | 0.865 | 32 | 0.6 | 0.30 |
| -0.468 | -0.440 | 0.738 | 0.262 | 0.872 | 33 | 0.6 | 0.30 |
| -0.440 | -0.413 | 0.737 | 0.263 | 0.878 | 34 | 0.6 | 0.30 |
| -0.413 | -0.386 | 0.735 | 0.265 | 0.885 | 35 | 0.6 | 0.30 |
| -0.386 | -0.359 | 0.733 | 0.267 | 0.891 | 36 | 0.6 | 0.30 |
| -0.359 | -0.332 | 0.731 | 0.269 | 0.897 | 37 | 0.6 | 0.30 |
| -0.332 | -0.306 | 0.729 | 0.271 | 0.904 | 38 | 0.6 | 0.30 |
| -0.306 | -0.280 | 0.727 | 0.273 | 0.910 | 39 | 0.6 | 0.30 |
| -0.280 | -0.254 | 0.725 | 0.275 | 0.916 | 40 | 0.6 | 0.30 |
| -0.254 | -0.228 | 0.723 | 0.277 | 0.923 | 41 | 0.6 | 0.30 |
| -0.228 | -0.203 | 0.721 | 0.279 | 0.929 | 42 | 0.6 | 0.30 |
| -0.203 | -0.177 | 0.719 | 0.281 | 0.935 | 43 | 0.6 | 0.30 |
| -0.177 | -0.152 | 0.718 | 0.282 | 0.941 | 44 | 0.6 | 0.30 |
| -0.152 | -0.126 | 0.716 | 0.284 | 0.948 | 45 | 0.6 | 0.30 |
| -0.126 | -0.101 | 0.714 | 0.286 | 0.954 | 46 | 0.6 | 0.30 |
| -0.101 | -0.076 | 0.712 | 0.288 | 0.960 | 47 | 0.6 | 0.30 |
| -0.076 | -0.051 | 0.710 | 0.290 | 0.966 | 48 | 0.6 | 0.30 |
| -0.051 | -0.026 | 0.708 | 0.292 | 0.973 | 49 | 0.6 | 0.30 |
| -0.026 | -0.001 | 0.706 | 0.294 | 0.979 | 50 | 0.6 | 0.30 |
| -0.001 | 0.024 | 0.704 | 0.296 | 0.985 | 51 | 0.6 | 0.30 |
| 0.024 | 0.050 | 0.703 | 0.297 | 0.991 | 52 | 0.6 | 0.30 |
| 0.050 | 0.075 | 0.701 | 0.299 | 0.998 | 53 | 0.6 | 0.30 |
| 0.075 | 0.100 | 0.699 | 0.301 | 1.004 | 54 | 0.6 | 0.30 |
| 0.100 | 0.125 | 0.697 | 0.303 | 1.011 | 55 | 0.6 | 0.30 |
| 0.125 | 0.150 | 0.695 | 0.305 | 1.017 | 56 | 0.6 | 0.30 |
| 0.150 | 0.176 | 0.693 | 0.307 | 1.024 | 57 | 0.6 | 0.30 |
| 0.176 | 0.201 | 0.691 | 0.309 | 1.030 | 58 | 0.6 | 0.30 |
| 0.201 | 0.227 | 0.689 | 0.311 | 1.037 | 59 | 0.6 | 0.30 |
| 0.227 | 0.253 | 0.687 | 0.313 | 1.044 | 60 | 0.6 | 0.30 |
| 0.253 | 0.279 | 0.685 | 0.315 | 1.050 | 61 | 0.6 | 0.30 |
| 0.279 | 0.305 | 0.683 | 0.317 | 1.057 | 62 | 0.6 | 0.30 |
| 0.305 | 0.331 | 0.681 | 0.319 | 1.064 | 63 | 0.6 | 0.30 |
| 0.331 | 0.358 | 0.679 | 0.321 | 1.071 | 64 | 0.6 | 0.30 |
| 0.358 | 0.385 | 0.677 | 0.323 | 1.078 | 65 | 0.6 | 0.30 |
| 0.385 | 0.412 | 0.674 | 0.326 | 1.085 | 66 | 0.6 | 0.30 |
| 0.412 | 0.439 | 0.672 | 0.328 | 1.092 | 67 | 0.6 | 0.30 |
| 0.439 | 0.467 | 0.670 | 0.330 | 1.100 | 68 | 0.6 | 0.30 |
| 0.467 | 0.495 | 0.668 | 0.332 | 1.107 | 69 | 0.6 | 0.30 |
| 0.495 | 0.524 | 0.666 | 0.334 | 1.115 | 70 | 0.6 | 0.30 |
| 0.524 | 0.553 | 0.663 | 0.337 | 1.123 | 71 | 0.6 | 0.30 |
| 0.553 | 0.582 | 0.661 | 0.339 | 1.131 | 72 | 0.6 | 0.30 |
| 0.582 | 0.612 | 0.658 | 0.342 | 1.139 | 73 | 0.6 | 0.30 |
| 0.612 | 0.643 | 0.656 | 0.344 | 1.147 | 74 | 0.6 | 0.30 |
| 0.643 | 0.674 | 0.653 | 0.347 | 1.155 | 75 | 0.6 | 0.30 |
| 0.674 | 0.706 | 0.651 | 0.349 | 1.164 | 76 | 0.6 | 0.30 |
| 0.706 | 0.739 | 0.648 | 0.352 | 1.173 | 77 | 0.6 | 0.30 |
| 0.739 | 0.772 | 0.645 | 0.355 | 1.182 | 78 | 0.6 | 0.30 |
| 0.772 | 0.806 | 0.643 | 0.357 | 1.191 | 79 | 0.6 | 0.30 |
| 0.806 | 0.842 | 0.640 | 0.360 | 1.201 | 80 | 0.6 | 0.30 |
| 0.842 | 0.878 | 0.637 | 0.363 | 1.211 | 81 | 0.6 | 0.30 |
| 0.878 | 0.915 | 0.634 | 0.366 | 1.222 | 82 | 0.6 | 0.30 |
| 0.915 | 0.954 | 0.630 | 0.370 | 1.232 | 83 | 0.6 | 0.30 |
| 0.954 | 0.995 | 0.627 | 0.373 | 1.243 | 84 | 0.6 | 0.30 |
| 0.995 | 1.037 | 0.623 | 0.377 | 1.255 | 85 | 0.6 | 0.30 |
| 1.037 | 1.081 | 0.620 | 0.380 | 1.267 | 86 | 0.6 | 0.30 |
| 1.081 | 1.127 | 0.616 | 0.384 | 1.280 | 87 | 0.6 | 0.30 |
| 1.127 | 1.175 | 0.612 | 0.388 | 1.294 | 88 | 0.6 | 0.30 |
| 1.175 | 1.227 | 0.608 | 0.392 | 1.308 | 89 | 0.6 | 0.30 |
| 1.227 | 1.282 | 0.603 | 0.397 | 1.324 | 90 | 0.6 | 0.30 |
| 1.282 | 1.341 | 0.598 | 0.402 | 1.340 | 91 | 0.6 | 0.30 |
| 1.341 | 1.406 | 0.593 | 0.407 | 1.358 | 92 | 0.6 | 0.30 |
| 1.406 | 1.477 | 0.587 | 0.413 | 1.378 | 93 | 0.6 | 0.30 |
| 1.477 | 1.556 | 0.580 | 0.420 | 1.400 | 94 | 0.6 | 0.30 |
| 1.556 | 1.646 | 0.573 | 0.427 | 1.425 | 95 | 0.6 | 0.30 |
| 1.646 | 1.752 | 0.564 | 0.436 | 1.454 | 96 | 0.6 | 0.30 |
| 1.752 | 1.882 | 0.553 | 0.447 | 1.489 | 97 | 0.6 | 0.30 |
| 1.882 | 2.056 | 0.540 | 0.460 | 1.534 | 98 | 0.6 | 0.30 |
| 2.056 | 2.329 | 0.520 | 0.480 | 1.598 | 99 | 0.6 | 0.30 |
| 2.329 | Inf | 0.476 | 0.524 | 1.746 | 100 | 0.6 | 0.30 |
| -Inf | -2.302 | 0.952 | 0.048 | 0.159 | 1 | 0.7 | 0.30 |
| -2.302 | -2.036 | 0.934 | 0.066 | 0.221 | 2 | 0.7 | 0.30 |
| -2.036 | -1.867 | 0.923 | 0.077 | 0.257 | 3 | 0.7 | 0.30 |
| -1.867 | -1.739 | 0.914 | 0.086 | 0.286 | 4 | 0.7 | 0.30 |
| -1.739 | -1.636 | 0.907 | 0.093 | 0.310 | 5 | 0.7 | 0.30 |
| -1.636 | -1.547 | 0.901 | 0.099 | 0.332 | 6 | 0.7 | 0.30 |
| -1.547 | -1.470 | 0.895 | 0.105 | 0.351 | 7 | 0.7 | 0.30 |
| -1.470 | -1.400 | 0.889 | 0.111 | 0.370 | 8 | 0.7 | 0.30 |
| -1.400 | -1.337 | 0.884 | 0.116 | 0.387 | 9 | 0.7 | 0.30 |
| -1.337 | -1.279 | 0.879 | 0.121 | 0.404 | 10 | 0.7 | 0.30 |
| -1.279 | -1.224 | 0.874 | 0.126 | 0.420 | 11 | 0.7 | 0.30 |
| -1.224 | -1.174 | 0.869 | 0.131 | 0.435 | 12 | 0.7 | 0.30 |
| -1.174 | -1.126 | 0.865 | 0.135 | 0.450 | 13 | 0.7 | 0.30 |
| -1.126 | -1.080 | 0.861 | 0.139 | 0.464 | 14 | 0.7 | 0.30 |
| -1.080 | -1.037 | 0.856 | 0.144 | 0.478 | 15 | 0.7 | 0.30 |
| -1.037 | -0.995 | 0.852 | 0.148 | 0.492 | 16 | 0.7 | 0.30 |
| -0.995 | -0.955 | 0.848 | 0.152 | 0.506 | 17 | 0.7 | 0.30 |
| -0.955 | -0.917 | 0.844 | 0.156 | 0.519 | 18 | 0.7 | 0.30 |
| -0.917 | -0.880 | 0.840 | 0.160 | 0.532 | 19 | 0.7 | 0.30 |
| -0.880 | -0.844 | 0.836 | 0.164 | 0.545 | 20 | 0.7 | 0.30 |
| -0.844 | -0.809 | 0.833 | 0.167 | 0.558 | 21 | 0.7 | 0.30 |
| -0.809 | -0.775 | 0.829 | 0.171 | 0.570 | 22 | 0.7 | 0.30 |
| -0.775 | -0.742 | 0.825 | 0.175 | 0.583 | 23 | 0.7 | 0.30 |
| -0.742 | -0.710 | 0.821 | 0.179 | 0.595 | 24 | 0.7 | 0.30 |
| -0.710 | -0.678 | 0.818 | 0.182 | 0.608 | 25 | 0.7 | 0.30 |
| -0.678 | -0.647 | 0.814 | 0.186 | 0.620 | 26 | 0.7 | 0.30 |
| -0.647 | -0.617 | 0.810 | 0.190 | 0.632 | 27 | 0.7 | 0.30 |
| -0.617 | -0.587 | 0.807 | 0.193 | 0.644 | 28 | 0.7 | 0.30 |
| -0.587 | -0.558 | 0.803 | 0.197 | 0.656 | 29 | 0.7 | 0.30 |
| -0.558 | -0.529 | 0.799 | 0.201 | 0.668 | 30 | 0.7 | 0.30 |
| -0.529 | -0.501 | 0.796 | 0.204 | 0.681 | 31 | 0.7 | 0.30 |
| -0.501 | -0.473 | 0.792 | 0.208 | 0.693 | 32 | 0.7 | 0.30 |
| -0.473 | -0.445 | 0.789 | 0.211 | 0.705 | 33 | 0.7 | 0.30 |
| -0.445 | -0.418 | 0.785 | 0.215 | 0.717 | 34 | 0.7 | 0.30 |
| -0.418 | -0.391 | 0.781 | 0.219 | 0.729 | 35 | 0.7 | 0.30 |
| -0.391 | -0.364 | 0.778 | 0.222 | 0.741 | 36 | 0.7 | 0.30 |
| -0.364 | -0.337 | 0.774 | 0.226 | 0.753 | 37 | 0.7 | 0.30 |
| -0.337 | -0.311 | 0.770 | 0.230 | 0.765 | 38 | 0.7 | 0.30 |
| -0.311 | -0.285 | 0.767 | 0.233 | 0.777 | 39 | 0.7 | 0.30 |
| -0.285 | -0.259 | 0.763 | 0.237 | 0.789 | 40 | 0.7 | 0.30 |
| -0.259 | -0.233 | 0.759 | 0.241 | 0.802 | 41 | 0.7 | 0.30 |
| -0.233 | -0.207 | 0.756 | 0.244 | 0.814 | 42 | 0.7 | 0.30 |
| -0.207 | -0.182 | 0.752 | 0.248 | 0.826 | 43 | 0.7 | 0.30 |
| -0.182 | -0.157 | 0.748 | 0.252 | 0.839 | 44 | 0.7 | 0.30 |
| -0.157 | -0.131 | 0.745 | 0.255 | 0.851 | 45 | 0.7 | 0.30 |
| -0.131 | -0.106 | 0.741 | 0.259 | 0.864 | 46 | 0.7 | 0.30 |
| -0.106 | -0.081 | 0.737 | 0.263 | 0.877 | 47 | 0.7 | 0.30 |
| -0.081 | -0.056 | 0.733 | 0.267 | 0.889 | 48 | 0.7 | 0.30 |
| -0.056 | -0.031 | 0.729 | 0.271 | 0.902 | 49 | 0.7 | 0.30 |
| -0.031 | -0.005 | 0.725 | 0.275 | 0.915 | 50 | 0.7 | 0.30 |
| -0.005 | 0.020 | 0.721 | 0.279 | 0.928 | 51 | 0.7 | 0.30 |
| 0.020 | 0.045 | 0.717 | 0.283 | 0.942 | 52 | 0.7 | 0.30 |
| 0.045 | 0.070 | 0.713 | 0.287 | 0.955 | 53 | 0.7 | 0.30 |
| 0.070 | 0.095 | 0.709 | 0.291 | 0.969 | 54 | 0.7 | 0.30 |
| 0.095 | 0.121 | 0.705 | 0.295 | 0.982 | 55 | 0.7 | 0.30 |
| 0.121 | 0.146 | 0.701 | 0.299 | 0.996 | 56 | 0.7 | 0.30 |
| 0.146 | 0.171 | 0.697 | 0.303 | 1.010 | 57 | 0.7 | 0.30 |
| 0.171 | 0.197 | 0.693 | 0.307 | 1.024 | 58 | 0.7 | 0.30 |
| 0.197 | 0.223 | 0.688 | 0.312 | 1.039 | 59 | 0.7 | 0.30 |
| 0.223 | 0.249 | 0.684 | 0.316 | 1.053 | 60 | 0.7 | 0.30 |
| 0.249 | 0.275 | 0.680 | 0.320 | 1.068 | 61 | 0.7 | 0.30 |
| 0.275 | 0.301 | 0.675 | 0.325 | 1.083 | 62 | 0.7 | 0.30 |
| 0.301 | 0.328 | 0.671 | 0.329 | 1.098 | 63 | 0.7 | 0.30 |
| 0.328 | 0.354 | 0.666 | 0.334 | 1.113 | 64 | 0.7 | 0.30 |
| 0.354 | 0.381 | 0.661 | 0.339 | 1.129 | 65 | 0.7 | 0.30 |
| 0.381 | 0.409 | 0.657 | 0.343 | 1.145 | 66 | 0.7 | 0.30 |
| 0.409 | 0.436 | 0.652 | 0.348 | 1.161 | 67 | 0.7 | 0.30 |
| 0.436 | 0.464 | 0.647 | 0.353 | 1.178 | 68 | 0.7 | 0.30 |
| 0.464 | 0.493 | 0.642 | 0.358 | 1.195 | 69 | 0.7 | 0.30 |
| 0.493 | 0.521 | 0.636 | 0.364 | 1.212 | 70 | 0.7 | 0.30 |
| 0.521 | 0.551 | 0.631 | 0.369 | 1.229 | 71 | 0.7 | 0.30 |
| 0.551 | 0.580 | 0.626 | 0.374 | 1.247 | 72 | 0.7 | 0.30 |
| 0.580 | 0.611 | 0.620 | 0.380 | 1.266 | 73 | 0.7 | 0.30 |
| 0.611 | 0.641 | 0.615 | 0.385 | 1.284 | 74 | 0.7 | 0.30 |
| 0.641 | 0.673 | 0.609 | 0.391 | 1.304 | 75 | 0.7 | 0.30 |
| 0.673 | 0.705 | 0.603 | 0.397 | 1.323 | 76 | 0.7 | 0.30 |
| 0.705 | 0.738 | 0.597 | 0.403 | 1.344 | 77 | 0.7 | 0.30 |
| 0.738 | 0.771 | 0.591 | 0.409 | 1.365 | 78 | 0.7 | 0.30 |
| 0.771 | 0.806 | 0.584 | 0.416 | 1.386 | 79 | 0.7 | 0.30 |
| 0.806 | 0.842 | 0.577 | 0.423 | 1.409 | 80 | 0.7 | 0.30 |
| 0.842 | 0.878 | 0.570 | 0.430 | 1.432 | 81 | 0.7 | 0.30 |
| 0.878 | 0.916 | 0.563 | 0.437 | 1.456 | 82 | 0.7 | 0.30 |
| 0.916 | 0.955 | 0.556 | 0.444 | 1.480 | 83 | 0.7 | 0.30 |
| 0.955 | 0.996 | 0.548 | 0.452 | 1.506 | 84 | 0.7 | 0.30 |
| 0.996 | 1.038 | 0.540 | 0.460 | 1.533 | 85 | 0.7 | 0.30 |
| 1.038 | 1.083 | 0.532 | 0.468 | 1.561 | 86 | 0.7 | 0.30 |
| 1.083 | 1.129 | 0.523 | 0.477 | 1.591 | 87 | 0.7 | 0.30 |
| 1.129 | 1.179 | 0.513 | 0.487 | 1.622 | 88 | 0.7 | 0.30 |
| 1.179 | 1.231 | 0.503 | 0.497 | 1.655 | 89 | 0.7 | 0.30 |
| 1.231 | 1.287 | 0.493 | 0.507 | 1.690 | 90 | 0.7 | 0.30 |
| 1.287 | 1.346 | 0.482 | 0.518 | 1.728 | 91 | 0.7 | 0.30 |
| 1.346 | 1.412 | 0.469 | 0.531 | 1.769 | 92 | 0.7 | 0.30 |
| 1.412 | 1.483 | 0.456 | 0.544 | 1.813 | 93 | 0.7 | 0.30 |
| 1.483 | 1.563 | 0.441 | 0.559 | 1.862 | 94 | 0.7 | 0.30 |
| 1.563 | 1.654 | 0.425 | 0.575 | 1.916 | 95 | 0.7 | 0.30 |
| 1.654 | 1.762 | 0.406 | 0.594 | 1.979 | 96 | 0.7 | 0.30 |
| 1.762 | 1.893 | 0.384 | 0.616 | 2.053 | 97 | 0.7 | 0.30 |
| 1.893 | 2.068 | 0.357 | 0.643 | 2.145 | 98 | 0.7 | 0.30 |
| 2.068 | 2.344 | 0.319 | 0.681 | 2.271 | 99 | 0.7 | 0.30 |
| 2.344 | Inf | 0.244 | 0.756 | 2.520 | 100 | 0.7 | 0.30 |
| -Inf | -2.239 | 0.989 | 0.011 | 0.036 | 1 | 0.8 | 0.30 |
| -2.239 | -1.989 | 0.982 | 0.018 | 0.061 | 2 | 0.8 | 0.30 |
| -1.989 | -1.829 | 0.976 | 0.024 | 0.080 | 3 | 0.8 | 0.30 |
| -1.829 | -1.709 | 0.971 | 0.029 | 0.095 | 4 | 0.8 | 0.30 |
| -1.709 | -1.610 | 0.967 | 0.033 | 0.110 | 5 | 0.8 | 0.30 |
| -1.610 | -1.526 | 0.963 | 0.037 | 0.124 | 6 | 0.8 | 0.30 |
| -1.526 | -1.452 | 0.959 | 0.041 | 0.137 | 7 | 0.8 | 0.30 |
| -1.452 | -1.386 | 0.955 | 0.045 | 0.150 | 8 | 0.8 | 0.30 |
| -1.386 | -1.326 | 0.951 | 0.049 | 0.163 | 9 | 0.8 | 0.30 |
| -1.326 | -1.270 | 0.947 | 0.053 | 0.176 | 10 | 0.8 | 0.30 |
| -1.270 | -1.218 | 0.944 | 0.056 | 0.188 | 11 | 0.8 | 0.30 |
| -1.218 | -1.169 | 0.940 | 0.060 | 0.201 | 12 | 0.8 | 0.30 |
| -1.169 | -1.123 | 0.936 | 0.064 | 0.213 | 13 | 0.8 | 0.30 |
| -1.123 | -1.079 | 0.932 | 0.068 | 0.226 | 14 | 0.8 | 0.30 |
| -1.079 | -1.037 | 0.929 | 0.071 | 0.238 | 15 | 0.8 | 0.30 |
| -1.037 | -0.997 | 0.925 | 0.075 | 0.251 | 16 | 0.8 | 0.30 |
| -0.997 | -0.959 | 0.921 | 0.079 | 0.263 | 17 | 0.8 | 0.30 |
| -0.959 | -0.921 | 0.917 | 0.083 | 0.276 | 18 | 0.8 | 0.30 |
| -0.921 | -0.885 | 0.913 | 0.087 | 0.289 | 19 | 0.8 | 0.30 |
| -0.885 | -0.850 | 0.909 | 0.091 | 0.302 | 20 | 0.8 | 0.30 |
| -0.850 | -0.817 | 0.906 | 0.094 | 0.315 | 21 | 0.8 | 0.30 |
| -0.817 | -0.783 | 0.902 | 0.098 | 0.328 | 22 | 0.8 | 0.30 |
| -0.783 | -0.751 | 0.898 | 0.102 | 0.341 | 23 | 0.8 | 0.30 |
| -0.751 | -0.720 | 0.894 | 0.106 | 0.355 | 24 | 0.8 | 0.30 |
| -0.720 | -0.689 | 0.889 | 0.111 | 0.368 | 25 | 0.8 | 0.30 |
| -0.689 | -0.659 | 0.885 | 0.115 | 0.382 | 26 | 0.8 | 0.30 |
| -0.659 | -0.629 | 0.881 | 0.119 | 0.396 | 27 | 0.8 | 0.30 |
| -0.629 | -0.600 | 0.877 | 0.123 | 0.410 | 28 | 0.8 | 0.30 |
| -0.600 | -0.571 | 0.873 | 0.127 | 0.425 | 29 | 0.8 | 0.30 |
| -0.571 | -0.542 | 0.868 | 0.132 | 0.439 | 30 | 0.8 | 0.30 |
| -0.542 | -0.514 | 0.864 | 0.136 | 0.454 | 31 | 0.8 | 0.30 |
| -0.514 | -0.487 | 0.859 | 0.141 | 0.469 | 32 | 0.8 | 0.30 |
| -0.487 | -0.460 | 0.855 | 0.145 | 0.484 | 33 | 0.8 | 0.30 |
| -0.460 | -0.432 | 0.850 | 0.150 | 0.500 | 34 | 0.8 | 0.30 |
| -0.432 | -0.406 | 0.845 | 0.155 | 0.515 | 35 | 0.8 | 0.30 |
| -0.406 | -0.379 | 0.841 | 0.159 | 0.531 | 36 | 0.8 | 0.30 |
| -0.379 | -0.353 | 0.836 | 0.164 | 0.548 | 37 | 0.8 | 0.30 |
| -0.353 | -0.327 | 0.831 | 0.169 | 0.564 | 38 | 0.8 | 0.30 |
| -0.327 | -0.301 | 0.826 | 0.174 | 0.581 | 39 | 0.8 | 0.30 |
| -0.301 | -0.275 | 0.821 | 0.179 | 0.598 | 40 | 0.8 | 0.30 |
| -0.275 | -0.249 | 0.815 | 0.185 | 0.615 | 41 | 0.8 | 0.30 |
| -0.249 | -0.224 | 0.810 | 0.190 | 0.633 | 42 | 0.8 | 0.30 |
| -0.224 | -0.198 | 0.805 | 0.195 | 0.651 | 43 | 0.8 | 0.30 |
| -0.198 | -0.173 | 0.799 | 0.201 | 0.669 | 44 | 0.8 | 0.30 |
| -0.173 | -0.147 | 0.794 | 0.206 | 0.687 | 45 | 0.8 | 0.30 |
| -0.147 | -0.122 | 0.788 | 0.212 | 0.706 | 46 | 0.8 | 0.30 |
| -0.122 | -0.097 | 0.782 | 0.218 | 0.726 | 47 | 0.8 | 0.30 |
| -0.097 | -0.072 | 0.776 | 0.224 | 0.745 | 48 | 0.8 | 0.30 |
| -0.072 | -0.046 | 0.770 | 0.230 | 0.765 | 49 | 0.8 | 0.30 |
| -0.046 | -0.021 | 0.764 | 0.236 | 0.786 | 50 | 0.8 | 0.30 |
| -0.021 | 0.004 | 0.758 | 0.242 | 0.806 | 51 | 0.8 | 0.30 |
| 0.004 | 0.030 | 0.752 | 0.248 | 0.828 | 52 | 0.8 | 0.30 |
| 0.030 | 0.055 | 0.745 | 0.255 | 0.849 | 53 | 0.8 | 0.30 |
| 0.055 | 0.080 | 0.739 | 0.261 | 0.871 | 54 | 0.8 | 0.30 |
| 0.080 | 0.106 | 0.732 | 0.268 | 0.894 | 55 | 0.8 | 0.30 |
| 0.106 | 0.132 | 0.725 | 0.275 | 0.917 | 56 | 0.8 | 0.30 |
| 0.132 | 0.158 | 0.718 | 0.282 | 0.940 | 57 | 0.8 | 0.30 |
| 0.158 | 0.184 | 0.711 | 0.289 | 0.964 | 58 | 0.8 | 0.30 |
| 0.184 | 0.210 | 0.703 | 0.297 | 0.988 | 59 | 0.8 | 0.30 |
| 0.210 | 0.236 | 0.696 | 0.304 | 1.013 | 60 | 0.8 | 0.30 |
| 0.236 | 0.263 | 0.688 | 0.312 | 1.039 | 61 | 0.8 | 0.30 |
| 0.263 | 0.290 | 0.681 | 0.319 | 1.065 | 62 | 0.8 | 0.30 |
| 0.290 | 0.317 | 0.673 | 0.327 | 1.092 | 63 | 0.8 | 0.30 |
| 0.317 | 0.344 | 0.664 | 0.336 | 1.119 | 64 | 0.8 | 0.30 |
| 0.344 | 0.372 | 0.656 | 0.344 | 1.147 | 65 | 0.8 | 0.30 |
| 0.372 | 0.400 | 0.647 | 0.353 | 1.175 | 66 | 0.8 | 0.30 |
| 0.400 | 0.428 | 0.639 | 0.361 | 1.204 | 67 | 0.8 | 0.30 |
| 0.428 | 0.457 | 0.630 | 0.370 | 1.234 | 68 | 0.8 | 0.30 |
| 0.457 | 0.486 | 0.621 | 0.379 | 1.265 | 69 | 0.8 | 0.30 |
| 0.486 | 0.515 | 0.611 | 0.389 | 1.296 | 70 | 0.8 | 0.30 |
| 0.515 | 0.545 | 0.602 | 0.398 | 1.328 | 71 | 0.8 | 0.30 |
| 0.545 | 0.576 | 0.592 | 0.408 | 1.361 | 72 | 0.8 | 0.30 |
| 0.576 | 0.607 | 0.582 | 0.418 | 1.395 | 73 | 0.8 | 0.30 |
| 0.607 | 0.639 | 0.571 | 0.429 | 1.430 | 74 | 0.8 | 0.30 |
| 0.639 | 0.671 | 0.560 | 0.440 | 1.465 | 75 | 0.8 | 0.30 |
| 0.671 | 0.704 | 0.550 | 0.450 | 1.502 | 76 | 0.8 | 0.30 |
| 0.704 | 0.738 | 0.538 | 0.462 | 1.539 | 77 | 0.8 | 0.30 |
| 0.738 | 0.773 | 0.527 | 0.473 | 1.578 | 78 | 0.8 | 0.30 |
| 0.773 | 0.808 | 0.515 | 0.485 | 1.617 | 79 | 0.8 | 0.30 |
| 0.808 | 0.845 | 0.503 | 0.497 | 1.658 | 80 | 0.8 | 0.30 |
| 0.845 | 0.883 | 0.490 | 0.510 | 1.700 | 81 | 0.8 | 0.30 |
| 0.883 | 0.922 | 0.477 | 0.523 | 1.744 | 82 | 0.8 | 0.30 |
| 0.922 | 0.962 | 0.463 | 0.537 | 1.789 | 83 | 0.8 | 0.30 |
| 0.962 | 1.005 | 0.450 | 0.550 | 1.835 | 84 | 0.8 | 0.30 |
| 1.005 | 1.048 | 0.435 | 0.565 | 1.883 | 85 | 0.8 | 0.30 |
| 1.048 | 1.094 | 0.420 | 0.580 | 1.932 | 86 | 0.8 | 0.30 |
| 1.094 | 1.142 | 0.405 | 0.595 | 1.984 | 87 | 0.8 | 0.30 |
| 1.142 | 1.193 | 0.389 | 0.611 | 2.037 | 88 | 0.8 | 0.30 |
| 1.193 | 1.247 | 0.372 | 0.628 | 2.093 | 89 | 0.8 | 0.30 |
| 1.247 | 1.304 | 0.355 | 0.645 | 2.151 | 90 | 0.8 | 0.30 |
| 1.304 | 1.366 | 0.337 | 0.663 | 2.212 | 91 | 0.8 | 0.30 |
| 1.366 | 1.433 | 0.317 | 0.683 | 2.275 | 92 | 0.8 | 0.30 |
| 1.433 | 1.506 | 0.297 | 0.703 | 2.343 | 93 | 0.8 | 0.30 |
| 1.506 | 1.588 | 0.276 | 0.724 | 2.414 | 94 | 0.8 | 0.30 |
| 1.588 | 1.681 | 0.253 | 0.747 | 2.491 | 95 | 0.8 | 0.30 |
| 1.681 | 1.790 | 0.228 | 0.772 | 2.573 | 96 | 0.8 | 0.30 |
| 1.790 | 1.923 | 0.201 | 0.799 | 2.664 | 97 | 0.8 | 0.30 |
| 1.923 | 2.099 | 0.170 | 0.830 | 2.768 | 98 | 0.8 | 0.30 |
| 2.099 | 2.373 | 0.133 | 0.867 | 2.892 | 99 | 0.8 | 0.30 |
| 2.373 | Inf | 0.077 | 0.923 | 3.075 | 100 | 0.8 | 0.30 |
| -Inf | -2.103 | 0.999 | 0.001 | 0.003 | 1 | 0.9 | 0.30 |
| -2.103 | -1.882 | 0.998 | 0.002 | 0.007 | 2 | 0.9 | 0.30 |
| -1.882 | -1.741 | 0.997 | 0.003 | 0.010 | 3 | 0.9 | 0.30 |
| -1.741 | -1.634 | 0.996 | 0.004 | 0.014 | 4 | 0.9 | 0.30 |
| -1.634 | -1.547 | 0.995 | 0.005 | 0.017 | 5 | 0.9 | 0.30 |
| -1.547 | -1.472 | 0.994 | 0.006 | 0.021 | 6 | 0.9 | 0.30 |
| -1.472 | -1.406 | 0.993 | 0.007 | 0.025 | 7 | 0.9 | 0.30 |
| -1.406 | -1.347 | 0.991 | 0.009 | 0.029 | 8 | 0.9 | 0.30 |
| -1.347 | -1.292 | 0.990 | 0.010 | 0.033 | 9 | 0.9 | 0.30 |
| -1.292 | -1.242 | 0.989 | 0.011 | 0.037 | 10 | 0.9 | 0.30 |
| -1.242 | -1.196 | 0.988 | 0.012 | 0.041 | 11 | 0.9 | 0.30 |
| -1.196 | -1.152 | 0.986 | 0.014 | 0.046 | 12 | 0.9 | 0.30 |
| -1.152 | -1.110 | 0.985 | 0.015 | 0.051 | 13 | 0.9 | 0.30 |
| -1.110 | -1.070 | 0.983 | 0.017 | 0.056 | 14 | 0.9 | 0.30 |
| -1.070 | -1.032 | 0.982 | 0.018 | 0.061 | 15 | 0.9 | 0.30 |
| -1.032 | -0.996 | 0.980 | 0.020 | 0.067 | 16 | 0.9 | 0.30 |
| -0.996 | -0.961 | 0.978 | 0.022 | 0.072 | 17 | 0.9 | 0.30 |
| -0.961 | -0.927 | 0.977 | 0.023 | 0.078 | 18 | 0.9 | 0.30 |
| -0.927 | -0.894 | 0.975 | 0.025 | 0.085 | 19 | 0.9 | 0.30 |
| -0.894 | -0.862 | 0.973 | 0.027 | 0.091 | 20 | 0.9 | 0.30 |
| -0.862 | -0.831 | 0.971 | 0.029 | 0.098 | 21 | 0.9 | 0.30 |
| -0.831 | -0.800 | 0.968 | 0.032 | 0.105 | 22 | 0.9 | 0.30 |
| -0.800 | -0.770 | 0.966 | 0.034 | 0.113 | 23 | 0.9 | 0.30 |
| -0.770 | -0.741 | 0.964 | 0.036 | 0.120 | 24 | 0.9 | 0.30 |
| -0.741 | -0.712 | 0.961 | 0.039 | 0.129 | 25 | 0.9 | 0.30 |
| -0.712 | -0.684 | 0.959 | 0.041 | 0.137 | 26 | 0.9 | 0.30 |
| -0.684 | -0.656 | 0.956 | 0.044 | 0.146 | 27 | 0.9 | 0.30 |
| -0.656 | -0.629 | 0.953 | 0.047 | 0.155 | 28 | 0.9 | 0.30 |
| -0.629 | -0.602 | 0.950 | 0.050 | 0.165 | 29 | 0.9 | 0.30 |
| -0.602 | -0.575 | 0.947 | 0.053 | 0.175 | 30 | 0.9 | 0.30 |
| -0.575 | -0.549 | 0.944 | 0.056 | 0.186 | 31 | 0.9 | 0.30 |
| -0.549 | -0.522 | 0.941 | 0.059 | 0.197 | 32 | 0.9 | 0.30 |
| -0.522 | -0.496 | 0.937 | 0.063 | 0.209 | 33 | 0.9 | 0.30 |
| -0.496 | -0.471 | 0.934 | 0.066 | 0.221 | 34 | 0.9 | 0.30 |
| -0.471 | -0.445 | 0.930 | 0.070 | 0.234 | 35 | 0.9 | 0.30 |
| -0.445 | -0.420 | 0.926 | 0.074 | 0.248 | 36 | 0.9 | 0.30 |
| -0.420 | -0.394 | 0.921 | 0.079 | 0.262 | 37 | 0.9 | 0.30 |
| -0.394 | -0.369 | 0.917 | 0.083 | 0.277 | 38 | 0.9 | 0.30 |
| -0.369 | -0.344 | 0.912 | 0.088 | 0.292 | 39 | 0.9 | 0.30 |
| -0.344 | -0.318 | 0.908 | 0.092 | 0.308 | 40 | 0.9 | 0.30 |
| -0.318 | -0.293 | 0.902 | 0.098 | 0.325 | 41 | 0.9 | 0.30 |
| -0.293 | -0.268 | 0.897 | 0.103 | 0.343 | 42 | 0.9 | 0.30 |
| -0.268 | -0.243 | 0.892 | 0.108 | 0.362 | 43 | 0.9 | 0.30 |
| -0.243 | -0.218 | 0.886 | 0.114 | 0.381 | 44 | 0.9 | 0.30 |
| -0.218 | -0.193 | 0.880 | 0.120 | 0.402 | 45 | 0.9 | 0.30 |
| -0.193 | -0.168 | 0.873 | 0.127 | 0.423 | 46 | 0.9 | 0.30 |
| -0.168 | -0.142 | 0.866 | 0.134 | 0.445 | 47 | 0.9 | 0.30 |
| -0.142 | -0.117 | 0.859 | 0.141 | 0.469 | 48 | 0.9 | 0.30 |
| -0.117 | -0.091 | 0.852 | 0.148 | 0.494 | 49 | 0.9 | 0.30 |
| -0.091 | -0.066 | 0.844 | 0.156 | 0.520 | 50 | 0.9 | 0.30 |
| -0.066 | -0.040 | 0.836 | 0.164 | 0.547 | 51 | 0.9 | 0.30 |
| -0.040 | -0.014 | 0.827 | 0.173 | 0.575 | 52 | 0.9 | 0.30 |
| -0.014 | 0.012 | 0.819 | 0.181 | 0.605 | 53 | 0.9 | 0.30 |
| 0.012 | 0.038 | 0.809 | 0.191 | 0.636 | 54 | 0.9 | 0.30 |
| 0.038 | 0.065 | 0.799 | 0.201 | 0.669 | 55 | 0.9 | 0.30 |
| 0.065 | 0.092 | 0.789 | 0.211 | 0.703 | 56 | 0.9 | 0.30 |
| 0.092 | 0.119 | 0.778 | 0.222 | 0.739 | 57 | 0.9 | 0.30 |
| 0.119 | 0.146 | 0.767 | 0.233 | 0.776 | 58 | 0.9 | 0.30 |
| 0.146 | 0.174 | 0.755 | 0.245 | 0.815 | 59 | 0.9 | 0.30 |
| 0.174 | 0.202 | 0.743 | 0.257 | 0.856 | 60 | 0.9 | 0.30 |
| 0.202 | 0.230 | 0.730 | 0.270 | 0.899 | 61 | 0.9 | 0.30 |
| 0.230 | 0.258 | 0.717 | 0.283 | 0.944 | 62 | 0.9 | 0.30 |
| 0.258 | 0.287 | 0.703 | 0.297 | 0.990 | 63 | 0.9 | 0.30 |
| 0.287 | 0.317 | 0.688 | 0.312 | 1.039 | 64 | 0.9 | 0.30 |
| 0.317 | 0.347 | 0.673 | 0.327 | 1.089 | 65 | 0.9 | 0.30 |
| 0.347 | 0.377 | 0.657 | 0.343 | 1.142 | 66 | 0.9 | 0.30 |
| 0.377 | 0.408 | 0.641 | 0.359 | 1.197 | 67 | 0.9 | 0.30 |
| 0.408 | 0.439 | 0.624 | 0.376 | 1.253 | 68 | 0.9 | 0.30 |
| 0.439 | 0.471 | 0.606 | 0.394 | 1.312 | 69 | 0.9 | 0.30 |
| 0.471 | 0.503 | 0.588 | 0.412 | 1.372 | 70 | 0.9 | 0.30 |
| 0.503 | 0.536 | 0.570 | 0.430 | 1.435 | 71 | 0.9 | 0.30 |
| 0.536 | 0.570 | 0.550 | 0.450 | 1.499 | 72 | 0.9 | 0.30 |
| 0.570 | 0.604 | 0.530 | 0.470 | 1.566 | 73 | 0.9 | 0.30 |
| 0.604 | 0.639 | 0.510 | 0.490 | 1.634 | 74 | 0.9 | 0.30 |
| 0.639 | 0.675 | 0.489 | 0.511 | 1.703 | 75 | 0.9 | 0.30 |
| 0.675 | 0.712 | 0.468 | 0.532 | 1.774 | 76 | 0.9 | 0.30 |
| 0.712 | 0.749 | 0.446 | 0.554 | 1.846 | 77 | 0.9 | 0.30 |
| 0.749 | 0.787 | 0.424 | 0.576 | 1.919 | 78 | 0.9 | 0.30 |
| 0.787 | 0.827 | 0.402 | 0.598 | 1.993 | 79 | 0.9 | 0.30 |
| 0.827 | 0.867 | 0.380 | 0.620 | 2.068 | 80 | 0.9 | 0.30 |
| 0.867 | 0.909 | 0.357 | 0.643 | 2.143 | 81 | 0.9 | 0.30 |
| 0.909 | 0.952 | 0.335 | 0.665 | 2.218 | 82 | 0.9 | 0.30 |
| 0.952 | 0.996 | 0.312 | 0.688 | 2.292 | 83 | 0.9 | 0.30 |
| 0.996 | 1.042 | 0.290 | 0.710 | 2.367 | 84 | 0.9 | 0.30 |
| 1.042 | 1.089 | 0.268 | 0.732 | 2.440 | 85 | 0.9 | 0.30 |
| 1.089 | 1.138 | 0.246 | 0.754 | 2.513 | 86 | 0.9 | 0.30 |
| 1.138 | 1.190 | 0.225 | 0.775 | 2.584 | 87 | 0.9 | 0.30 |
| 1.190 | 1.243 | 0.204 | 0.796 | 2.653 | 88 | 0.9 | 0.30 |
| 1.243 | 1.300 | 0.184 | 0.816 | 2.720 | 89 | 0.9 | 0.30 |
| 1.300 | 1.360 | 0.164 | 0.836 | 2.786 | 90 | 0.9 | 0.30 |
| 1.360 | 1.423 | 0.145 | 0.855 | 2.849 | 91 | 0.9 | 0.30 |
| 1.423 | 1.491 | 0.127 | 0.873 | 2.910 | 92 | 0.9 | 0.30 |
| 1.491 | 1.565 | 0.110 | 0.890 | 2.968 | 93 | 0.9 | 0.30 |
| 1.565 | 1.647 | 0.093 | 0.907 | 3.023 | 94 | 0.9 | 0.30 |
| 1.647 | 1.739 | 0.077 | 0.923 | 3.076 | 95 | 0.9 | 0.30 |
| 1.739 | 1.844 | 0.062 | 0.938 | 3.125 | 96 | 0.9 | 0.30 |
| 1.844 | 1.972 | 0.048 | 0.952 | 3.173 | 97 | 0.9 | 0.30 |
| 1.972 | 2.137 | 0.035 | 0.965 | 3.217 | 98 | 0.9 | 0.30 |
| 2.137 | 2.390 | 0.022 | 0.978 | 3.259 | 99 | 0.9 | 0.30 |
| 2.390 | Inf | 0.009 | 0.991 | 3.303 | 100 | 0.9 | 0.30 |
| -Inf | -2.326 | 0.724 | 0.276 | 0.551 | 1 | 0.6 | 0.50 |
| -2.326 | -2.054 | 0.688 | 0.312 | 0.623 | 2 | 0.6 | 0.50 |
| -2.054 | -1.881 | 0.671 | 0.329 | 0.657 | 3 | 0.6 | 0.50 |
| -1.881 | -1.751 | 0.659 | 0.341 | 0.682 | 4 | 0.6 | 0.50 |
| -1.751 | -1.645 | 0.650 | 0.350 | 0.701 | 5 | 0.6 | 0.50 |
| -1.645 | -1.555 | 0.642 | 0.358 | 0.717 | 6 | 0.6 | 0.50 |
| -1.555 | -1.476 | 0.634 | 0.366 | 0.731 | 7 | 0.6 | 0.50 |
| -1.476 | -1.405 | 0.628 | 0.372 | 0.744 | 8 | 0.6 | 0.50 |
| -1.405 | -1.341 | 0.622 | 0.378 | 0.755 | 9 | 0.6 | 0.50 |
| -1.341 | -1.282 | 0.617 | 0.383 | 0.766 | 10 | 0.6 | 0.50 |
| -1.282 | -1.227 | 0.612 | 0.388 | 0.776 | 11 | 0.6 | 0.50 |
| -1.227 | -1.175 | 0.608 | 0.392 | 0.785 | 12 | 0.6 | 0.50 |
| -1.175 | -1.127 | 0.603 | 0.397 | 0.794 | 13 | 0.6 | 0.50 |
| -1.127 | -1.080 | 0.599 | 0.401 | 0.802 | 14 | 0.6 | 0.50 |
| -1.080 | -1.037 | 0.595 | 0.405 | 0.810 | 15 | 0.6 | 0.50 |
| -1.037 | -0.995 | 0.591 | 0.409 | 0.817 | 16 | 0.6 | 0.50 |
| -0.995 | -0.954 | 0.588 | 0.412 | 0.825 | 17 | 0.6 | 0.50 |
| -0.954 | -0.916 | 0.584 | 0.416 | 0.831 | 18 | 0.6 | 0.50 |
| -0.916 | -0.878 | 0.581 | 0.419 | 0.838 | 19 | 0.6 | 0.50 |
| -0.878 | -0.842 | 0.578 | 0.422 | 0.845 | 20 | 0.6 | 0.50 |
| -0.842 | -0.807 | 0.574 | 0.426 | 0.851 | 21 | 0.6 | 0.50 |
| -0.807 | -0.772 | 0.571 | 0.429 | 0.857 | 22 | 0.6 | 0.50 |
| -0.772 | -0.739 | 0.568 | 0.432 | 0.863 | 23 | 0.6 | 0.50 |
| -0.739 | -0.706 | 0.565 | 0.435 | 0.869 | 24 | 0.6 | 0.50 |
| -0.706 | -0.675 | 0.563 | 0.437 | 0.875 | 25 | 0.6 | 0.50 |
| -0.675 | -0.643 | 0.560 | 0.440 | 0.881 | 26 | 0.6 | 0.50 |
| -0.643 | -0.613 | 0.557 | 0.443 | 0.886 | 27 | 0.6 | 0.50 |
| -0.613 | -0.583 | 0.554 | 0.446 | 0.892 | 28 | 0.6 | 0.50 |
| -0.583 | -0.554 | 0.552 | 0.448 | 0.897 | 29 | 0.6 | 0.50 |
| -0.554 | -0.525 | 0.549 | 0.451 | 0.902 | 30 | 0.6 | 0.50 |
| -0.525 | -0.496 | 0.546 | 0.454 | 0.907 | 31 | 0.6 | 0.50 |
| -0.496 | -0.468 | 0.544 | 0.456 | 0.913 | 32 | 0.6 | 0.50 |
| -0.468 | -0.440 | 0.541 | 0.459 | 0.918 | 33 | 0.6 | 0.50 |
| -0.440 | -0.413 | 0.539 | 0.461 | 0.923 | 34 | 0.6 | 0.50 |
| -0.413 | -0.385 | 0.536 | 0.464 | 0.928 | 35 | 0.6 | 0.50 |
| -0.385 | -0.359 | 0.534 | 0.466 | 0.932 | 36 | 0.6 | 0.50 |
| -0.359 | -0.332 | 0.531 | 0.469 | 0.937 | 37 | 0.6 | 0.50 |
| -0.332 | -0.306 | 0.529 | 0.471 | 0.942 | 38 | 0.6 | 0.50 |
| -0.306 | -0.279 | 0.527 | 0.473 | 0.947 | 39 | 0.6 | 0.50 |
| -0.279 | -0.253 | 0.524 | 0.476 | 0.952 | 40 | 0.6 | 0.50 |
| -0.253 | -0.228 | 0.522 | 0.478 | 0.956 | 41 | 0.6 | 0.50 |
| -0.228 | -0.202 | 0.520 | 0.480 | 0.961 | 42 | 0.6 | 0.50 |
| -0.202 | -0.176 | 0.517 | 0.483 | 0.966 | 43 | 0.6 | 0.50 |
| -0.176 | -0.151 | 0.515 | 0.485 | 0.970 | 44 | 0.6 | 0.50 |
| -0.151 | -0.126 | 0.513 | 0.487 | 0.975 | 45 | 0.6 | 0.50 |
| -0.126 | -0.100 | 0.510 | 0.490 | 0.979 | 46 | 0.6 | 0.50 |
| -0.100 | -0.075 | 0.508 | 0.492 | 0.984 | 47 | 0.6 | 0.50 |
| -0.075 | -0.050 | 0.506 | 0.494 | 0.989 | 48 | 0.6 | 0.50 |
| -0.050 | -0.025 | 0.503 | 0.497 | 0.993 | 49 | 0.6 | 0.50 |
| -0.025 | 0.000 | 0.501 | 0.499 | 0.998 | 50 | 0.6 | 0.50 |
| 0.000 | 0.025 | 0.499 | 0.501 | 1.002 | 51 | 0.6 | 0.50 |
| 0.025 | 0.050 | 0.497 | 0.503 | 1.007 | 52 | 0.6 | 0.50 |
| 0.050 | 0.075 | 0.494 | 0.506 | 1.011 | 53 | 0.6 | 0.50 |
| 0.075 | 0.100 | 0.492 | 0.508 | 1.016 | 54 | 0.6 | 0.50 |
| 0.100 | 0.126 | 0.490 | 0.510 | 1.021 | 55 | 0.6 | 0.50 |
| 0.126 | 0.151 | 0.487 | 0.513 | 1.025 | 56 | 0.6 | 0.50 |
| 0.151 | 0.176 | 0.485 | 0.515 | 1.030 | 57 | 0.6 | 0.50 |
| 0.176 | 0.202 | 0.483 | 0.517 | 1.034 | 58 | 0.6 | 0.50 |
| 0.202 | 0.228 | 0.480 | 0.520 | 1.039 | 59 | 0.6 | 0.50 |
| 0.228 | 0.253 | 0.478 | 0.522 | 1.044 | 60 | 0.6 | 0.50 |
| 0.253 | 0.279 | 0.476 | 0.524 | 1.048 | 61 | 0.6 | 0.50 |
| 0.279 | 0.306 | 0.473 | 0.527 | 1.053 | 62 | 0.6 | 0.50 |
| 0.306 | 0.332 | 0.471 | 0.529 | 1.058 | 63 | 0.6 | 0.50 |
| 0.332 | 0.359 | 0.469 | 0.531 | 1.063 | 64 | 0.6 | 0.50 |
| 0.359 | 0.385 | 0.466 | 0.534 | 1.068 | 65 | 0.6 | 0.50 |
| 0.385 | 0.413 | 0.464 | 0.536 | 1.072 | 66 | 0.6 | 0.50 |
| 0.413 | 0.440 | 0.461 | 0.539 | 1.077 | 67 | 0.6 | 0.50 |
| 0.440 | 0.468 | 0.459 | 0.541 | 1.082 | 68 | 0.6 | 0.50 |
| 0.468 | 0.496 | 0.456 | 0.544 | 1.087 | 69 | 0.6 | 0.50 |
| 0.496 | 0.525 | 0.454 | 0.546 | 1.093 | 70 | 0.6 | 0.50 |
| 0.525 | 0.554 | 0.451 | 0.549 | 1.098 | 71 | 0.6 | 0.50 |
| 0.554 | 0.583 | 0.448 | 0.552 | 1.103 | 72 | 0.6 | 0.50 |
| 0.583 | 0.613 | 0.446 | 0.554 | 1.108 | 73 | 0.6 | 0.50 |
| 0.613 | 0.643 | 0.443 | 0.557 | 1.114 | 74 | 0.6 | 0.50 |
| 0.643 | 0.675 | 0.440 | 0.560 | 1.119 | 75 | 0.6 | 0.50 |
| 0.675 | 0.706 | 0.437 | 0.563 | 1.125 | 76 | 0.6 | 0.50 |
| 0.706 | 0.739 | 0.435 | 0.565 | 1.131 | 77 | 0.6 | 0.50 |
| 0.739 | 0.772 | 0.432 | 0.568 | 1.137 | 78 | 0.6 | 0.50 |
| 0.772 | 0.807 | 0.429 | 0.571 | 1.143 | 79 | 0.6 | 0.50 |
| 0.807 | 0.842 | 0.426 | 0.574 | 1.149 | 80 | 0.6 | 0.50 |
| 0.842 | 0.878 | 0.422 | 0.578 | 1.155 | 81 | 0.6 | 0.50 |
| 0.878 | 0.916 | 0.419 | 0.581 | 1.162 | 82 | 0.6 | 0.50 |
| 0.916 | 0.954 | 0.416 | 0.584 | 1.169 | 83 | 0.6 | 0.50 |
| 0.954 | 0.995 | 0.412 | 0.588 | 1.175 | 84 | 0.6 | 0.50 |
| 0.995 | 1.037 | 0.409 | 0.591 | 1.183 | 85 | 0.6 | 0.50 |
| 1.037 | 1.080 | 0.405 | 0.595 | 1.190 | 86 | 0.6 | 0.50 |
| 1.080 | 1.127 | 0.401 | 0.599 | 1.198 | 87 | 0.6 | 0.50 |
| 1.127 | 1.175 | 0.397 | 0.603 | 1.206 | 88 | 0.6 | 0.50 |
| 1.175 | 1.227 | 0.392 | 0.608 | 1.215 | 89 | 0.6 | 0.50 |
| 1.227 | 1.282 | 0.388 | 0.612 | 1.224 | 90 | 0.6 | 0.50 |
| 1.282 | 1.341 | 0.383 | 0.617 | 1.234 | 91 | 0.6 | 0.50 |
| 1.341 | 1.405 | 0.378 | 0.622 | 1.245 | 92 | 0.6 | 0.50 |
| 1.405 | 1.476 | 0.372 | 0.628 | 1.256 | 93 | 0.6 | 0.50 |
| 1.476 | 1.555 | 0.366 | 0.634 | 1.269 | 94 | 0.6 | 0.50 |
| 1.555 | 1.645 | 0.358 | 0.642 | 1.283 | 95 | 0.6 | 0.50 |
| 1.645 | 1.751 | 0.350 | 0.650 | 1.299 | 96 | 0.6 | 0.50 |
| 1.751 | 1.881 | 0.341 | 0.659 | 1.318 | 97 | 0.6 | 0.50 |
| 1.881 | 2.054 | 0.329 | 0.671 | 1.343 | 98 | 0.6 | 0.50 |
| 2.054 | 2.326 | 0.312 | 0.688 | 1.377 | 99 | 0.6 | 0.50 |
| 2.326 | Inf | 0.276 | 0.724 | 1.449 | 100 | 0.6 | 0.50 |
| -Inf | -2.319 | 0.889 | 0.111 | 0.223 | 1 | 0.7 | 0.50 |
| -2.319 | -2.050 | 0.848 | 0.152 | 0.305 | 2 | 0.7 | 0.50 |
| -2.050 | -1.879 | 0.825 | 0.175 | 0.350 | 3 | 0.7 | 0.50 |
| -1.879 | -1.750 | 0.807 | 0.193 | 0.385 | 4 | 0.7 | 0.50 |
| -1.750 | -1.645 | 0.793 | 0.207 | 0.415 | 5 | 0.7 | 0.50 |
| -1.645 | -1.556 | 0.780 | 0.220 | 0.440 | 6 | 0.7 | 0.50 |
| -1.556 | -1.477 | 0.768 | 0.232 | 0.463 | 7 | 0.7 | 0.50 |
| -1.477 | -1.407 | 0.758 | 0.242 | 0.485 | 8 | 0.7 | 0.50 |
| -1.407 | -1.343 | 0.748 | 0.252 | 0.504 | 9 | 0.7 | 0.50 |
| -1.343 | -1.284 | 0.739 | 0.261 | 0.523 | 10 | 0.7 | 0.50 |
| -1.284 | -1.229 | 0.730 | 0.270 | 0.540 | 11 | 0.7 | 0.50 |
| -1.229 | -1.177 | 0.721 | 0.279 | 0.557 | 12 | 0.7 | 0.50 |
| -1.177 | -1.129 | 0.713 | 0.287 | 0.573 | 13 | 0.7 | 0.50 |
| -1.129 | -1.083 | 0.706 | 0.294 | 0.589 | 14 | 0.7 | 0.50 |
| -1.083 | -1.039 | 0.698 | 0.302 | 0.603 | 15 | 0.7 | 0.50 |
| -1.039 | -0.997 | 0.691 | 0.309 | 0.618 | 16 | 0.7 | 0.50 |
| -0.997 | -0.957 | 0.684 | 0.316 | 0.632 | 17 | 0.7 | 0.50 |
| -0.957 | -0.918 | 0.677 | 0.323 | 0.645 | 18 | 0.7 | 0.50 |
| -0.918 | -0.881 | 0.671 | 0.329 | 0.659 | 19 | 0.7 | 0.50 |
| -0.881 | -0.844 | 0.664 | 0.336 | 0.672 | 20 | 0.7 | 0.50 |
| -0.844 | -0.809 | 0.658 | 0.342 | 0.684 | 21 | 0.7 | 0.50 |
| -0.809 | -0.775 | 0.652 | 0.348 | 0.697 | 22 | 0.7 | 0.50 |
| -0.775 | -0.741 | 0.646 | 0.354 | 0.709 | 23 | 0.7 | 0.50 |
| -0.741 | -0.709 | 0.640 | 0.360 | 0.721 | 24 | 0.7 | 0.50 |
| -0.709 | -0.677 | 0.634 | 0.366 | 0.733 | 25 | 0.7 | 0.50 |
| -0.677 | -0.646 | 0.628 | 0.372 | 0.744 | 26 | 0.7 | 0.50 |
| -0.646 | -0.615 | 0.622 | 0.378 | 0.756 | 27 | 0.7 | 0.50 |
| -0.615 | -0.585 | 0.616 | 0.384 | 0.767 | 28 | 0.7 | 0.50 |
| -0.585 | -0.555 | 0.611 | 0.389 | 0.778 | 29 | 0.7 | 0.50 |
| -0.555 | -0.526 | 0.605 | 0.395 | 0.789 | 30 | 0.7 | 0.50 |
| -0.526 | -0.498 | 0.600 | 0.400 | 0.800 | 31 | 0.7 | 0.50 |
| -0.498 | -0.470 | 0.594 | 0.406 | 0.811 | 32 | 0.7 | 0.50 |
| -0.470 | -0.442 | 0.589 | 0.411 | 0.822 | 33 | 0.7 | 0.50 |
| -0.442 | -0.414 | 0.584 | 0.416 | 0.832 | 34 | 0.7 | 0.50 |
| -0.414 | -0.387 | 0.579 | 0.421 | 0.843 | 35 | 0.7 | 0.50 |
| -0.387 | -0.360 | 0.573 | 0.427 | 0.853 | 36 | 0.7 | 0.50 |
| -0.360 | -0.333 | 0.568 | 0.432 | 0.864 | 37 | 0.7 | 0.50 |
| -0.333 | -0.307 | 0.563 | 0.437 | 0.874 | 38 | 0.7 | 0.50 |
| -0.307 | -0.280 | 0.558 | 0.442 | 0.884 | 39 | 0.7 | 0.50 |
| -0.280 | -0.254 | 0.553 | 0.447 | 0.895 | 40 | 0.7 | 0.50 |
| -0.254 | -0.229 | 0.548 | 0.452 | 0.905 | 41 | 0.7 | 0.50 |
| -0.229 | -0.203 | 0.543 | 0.457 | 0.915 | 42 | 0.7 | 0.50 |
| -0.203 | -0.177 | 0.537 | 0.463 | 0.925 | 43 | 0.7 | 0.50 |
| -0.177 | -0.152 | 0.532 | 0.468 | 0.935 | 44 | 0.7 | 0.50 |
| -0.152 | -0.126 | 0.527 | 0.473 | 0.945 | 45 | 0.7 | 0.50 |
| -0.126 | -0.101 | 0.522 | 0.478 | 0.955 | 46 | 0.7 | 0.50 |
| -0.101 | -0.076 | 0.517 | 0.483 | 0.965 | 47 | 0.7 | 0.50 |
| -0.076 | -0.050 | 0.512 | 0.488 | 0.975 | 48 | 0.7 | 0.50 |
| -0.050 | -0.025 | 0.507 | 0.493 | 0.985 | 49 | 0.7 | 0.50 |
| -0.025 | 0.000 | 0.502 | 0.498 | 0.995 | 50 | 0.7 | 0.50 |
| 0.000 | 0.025 | 0.498 | 0.502 | 1.005 | 51 | 0.7 | 0.50 |
| 0.025 | 0.050 | 0.493 | 0.507 | 1.015 | 52 | 0.7 | 0.50 |
| 0.050 | 0.076 | 0.488 | 0.512 | 1.025 | 53 | 0.7 | 0.50 |
| 0.076 | 0.101 | 0.483 | 0.517 | 1.035 | 54 | 0.7 | 0.50 |
| 0.101 | 0.126 | 0.478 | 0.522 | 1.045 | 55 | 0.7 | 0.50 |
| 0.126 | 0.152 | 0.473 | 0.527 | 1.055 | 56 | 0.7 | 0.50 |
| 0.152 | 0.177 | 0.468 | 0.532 | 1.065 | 57 | 0.7 | 0.50 |
| 0.177 | 0.203 | 0.463 | 0.537 | 1.075 | 58 | 0.7 | 0.50 |
| 0.203 | 0.229 | 0.457 | 0.543 | 1.085 | 59 | 0.7 | 0.50 |
| 0.229 | 0.254 | 0.452 | 0.548 | 1.095 | 60 | 0.7 | 0.50 |
| 0.254 | 0.280 | 0.447 | 0.553 | 1.105 | 61 | 0.7 | 0.50 |
| 0.280 | 0.307 | 0.442 | 0.558 | 1.116 | 62 | 0.7 | 0.50 |
| 0.307 | 0.333 | 0.437 | 0.563 | 1.126 | 63 | 0.7 | 0.50 |
| 0.333 | 0.360 | 0.432 | 0.568 | 1.136 | 64 | 0.7 | 0.50 |
| 0.360 | 0.387 | 0.427 | 0.573 | 1.147 | 65 | 0.7 | 0.50 |
| 0.387 | 0.414 | 0.421 | 0.579 | 1.157 | 66 | 0.7 | 0.50 |
| 0.414 | 0.442 | 0.416 | 0.584 | 1.168 | 67 | 0.7 | 0.50 |
| 0.442 | 0.470 | 0.411 | 0.589 | 1.178 | 68 | 0.7 | 0.50 |
| 0.470 | 0.498 | 0.406 | 0.594 | 1.189 | 69 | 0.7 | 0.50 |
| 0.498 | 0.526 | 0.400 | 0.600 | 1.200 | 70 | 0.7 | 0.50 |
| 0.526 | 0.555 | 0.395 | 0.605 | 1.211 | 71 | 0.7 | 0.50 |
| 0.555 | 0.585 | 0.389 | 0.611 | 1.222 | 72 | 0.7 | 0.50 |
| 0.585 | 0.615 | 0.384 | 0.616 | 1.233 | 73 | 0.7 | 0.50 |
| 0.615 | 0.646 | 0.378 | 0.622 | 1.244 | 74 | 0.7 | 0.50 |
| 0.646 | 0.677 | 0.372 | 0.628 | 1.256 | 75 | 0.7 | 0.50 |
| 0.677 | 0.709 | 0.366 | 0.634 | 1.267 | 76 | 0.7 | 0.50 |
| 0.709 | 0.741 | 0.360 | 0.640 | 1.279 | 77 | 0.7 | 0.50 |
| 0.741 | 0.775 | 0.354 | 0.646 | 1.291 | 78 | 0.7 | 0.50 |
| 0.775 | 0.809 | 0.348 | 0.652 | 1.303 | 79 | 0.7 | 0.50 |
| 0.809 | 0.844 | 0.342 | 0.658 | 1.316 | 80 | 0.7 | 0.50 |
| 0.844 | 0.881 | 0.336 | 0.664 | 1.328 | 81 | 0.7 | 0.50 |
| 0.881 | 0.918 | 0.329 | 0.671 | 1.341 | 82 | 0.7 | 0.50 |
| 0.918 | 0.957 | 0.323 | 0.677 | 1.355 | 83 | 0.7 | 0.50 |
| 0.957 | 0.997 | 0.316 | 0.684 | 1.368 | 84 | 0.7 | 0.50 |
| 0.997 | 1.039 | 0.309 | 0.691 | 1.382 | 85 | 0.7 | 0.50 |
| 1.039 | 1.083 | 0.302 | 0.698 | 1.397 | 86 | 0.7 | 0.50 |
| 1.083 | 1.129 | 0.294 | 0.706 | 1.411 | 87 | 0.7 | 0.50 |
| 1.129 | 1.177 | 0.287 | 0.713 | 1.427 | 88 | 0.7 | 0.50 |
| 1.177 | 1.229 | 0.279 | 0.721 | 1.443 | 89 | 0.7 | 0.50 |
| 1.229 | 1.284 | 0.270 | 0.730 | 1.460 | 90 | 0.7 | 0.50 |
| 1.284 | 1.343 | 0.261 | 0.739 | 1.477 | 91 | 0.7 | 0.50 |
| 1.343 | 1.407 | 0.252 | 0.748 | 1.496 | 92 | 0.7 | 0.50 |
| 1.407 | 1.477 | 0.242 | 0.758 | 1.515 | 93 | 0.7 | 0.50 |
| 1.477 | 1.556 | 0.232 | 0.768 | 1.537 | 94 | 0.7 | 0.50 |
| 1.556 | 1.645 | 0.220 | 0.780 | 1.560 | 95 | 0.7 | 0.50 |
| 1.645 | 1.750 | 0.207 | 0.793 | 1.585 | 96 | 0.7 | 0.50 |
| 1.750 | 1.879 | 0.193 | 0.807 | 1.615 | 97 | 0.7 | 0.50 |
| 1.879 | 2.050 | 0.175 | 0.825 | 1.650 | 98 | 0.7 | 0.50 |
| 2.050 | 2.319 | 0.152 | 0.848 | 1.695 | 99 | 0.7 | 0.50 |
| 2.319 | Inf | 0.111 | 0.889 | 1.777 | 100 | 0.7 | 0.50 |
| -Inf | -2.286 | 0.972 | 0.028 | 0.057 | 1 | 0.8 | 0.50 |
| -2.286 | -2.031 | 0.951 | 0.049 | 0.098 | 2 | 0.8 | 0.50 |
| -2.031 | -1.868 | 0.937 | 0.063 | 0.127 | 3 | 0.8 | 0.50 |
| -1.868 | -1.744 | 0.924 | 0.076 | 0.152 | 4 | 0.8 | 0.50 |
| -1.744 | -1.643 | 0.912 | 0.088 | 0.175 | 5 | 0.8 | 0.50 |
| -1.643 | -1.557 | 0.902 | 0.098 | 0.197 | 6 | 0.8 | 0.50 |
| -1.557 | -1.480 | 0.891 | 0.109 | 0.218 | 7 | 0.8 | 0.50 |
| -1.480 | -1.412 | 0.881 | 0.119 | 0.238 | 8 | 0.8 | 0.50 |
| -1.412 | -1.349 | 0.871 | 0.129 | 0.258 | 9 | 0.8 | 0.50 |
| -1.349 | -1.291 | 0.862 | 0.138 | 0.277 | 10 | 0.8 | 0.50 |
| -1.291 | -1.237 | 0.852 | 0.148 | 0.296 | 11 | 0.8 | 0.50 |
| -1.237 | -1.187 | 0.843 | 0.157 | 0.315 | 12 | 0.8 | 0.50 |
| -1.187 | -1.139 | 0.833 | 0.167 | 0.333 | 13 | 0.8 | 0.50 |
| -1.139 | -1.093 | 0.824 | 0.176 | 0.351 | 14 | 0.8 | 0.50 |
| -1.093 | -1.050 | 0.815 | 0.185 | 0.370 | 15 | 0.8 | 0.50 |
| -1.050 | -1.008 | 0.806 | 0.194 | 0.388 | 16 | 0.8 | 0.50 |
| -1.008 | -0.968 | 0.797 | 0.203 | 0.406 | 17 | 0.8 | 0.50 |
| -0.968 | -0.930 | 0.788 | 0.212 | 0.424 | 18 | 0.8 | 0.50 |
| -0.930 | -0.892 | 0.779 | 0.221 | 0.441 | 19 | 0.8 | 0.50 |
| -0.892 | -0.856 | 0.770 | 0.230 | 0.459 | 20 | 0.8 | 0.50 |
| -0.856 | -0.821 | 0.762 | 0.238 | 0.477 | 21 | 0.8 | 0.50 |
| -0.821 | -0.786 | 0.753 | 0.247 | 0.495 | 22 | 0.8 | 0.50 |
| -0.786 | -0.753 | 0.744 | 0.256 | 0.512 | 23 | 0.8 | 0.50 |
| -0.753 | -0.720 | 0.735 | 0.265 | 0.530 | 24 | 0.8 | 0.50 |
| -0.720 | -0.688 | 0.726 | 0.274 | 0.548 | 25 | 0.8 | 0.50 |
| -0.688 | -0.657 | 0.717 | 0.283 | 0.565 | 26 | 0.8 | 0.50 |
| -0.657 | -0.626 | 0.708 | 0.292 | 0.583 | 27 | 0.8 | 0.50 |
| -0.626 | -0.595 | 0.700 | 0.300 | 0.601 | 28 | 0.8 | 0.50 |
| -0.595 | -0.565 | 0.691 | 0.309 | 0.618 | 29 | 0.8 | 0.50 |
| -0.565 | -0.536 | 0.682 | 0.318 | 0.636 | 30 | 0.8 | 0.50 |
| -0.536 | -0.507 | 0.673 | 0.327 | 0.654 | 31 | 0.8 | 0.50 |
| -0.507 | -0.478 | 0.664 | 0.336 | 0.671 | 32 | 0.8 | 0.50 |
| -0.478 | -0.450 | 0.655 | 0.345 | 0.689 | 33 | 0.8 | 0.50 |
| -0.450 | -0.422 | 0.647 | 0.353 | 0.707 | 34 | 0.8 | 0.50 |
| -0.422 | -0.395 | 0.638 | 0.362 | 0.725 | 35 | 0.8 | 0.50 |
| -0.395 | -0.367 | 0.629 | 0.371 | 0.742 | 36 | 0.8 | 0.50 |
| -0.367 | -0.340 | 0.620 | 0.380 | 0.760 | 37 | 0.8 | 0.50 |
| -0.340 | -0.313 | 0.611 | 0.389 | 0.778 | 38 | 0.8 | 0.50 |
| -0.313 | -0.286 | 0.602 | 0.398 | 0.795 | 39 | 0.8 | 0.50 |
| -0.286 | -0.260 | 0.593 | 0.407 | 0.813 | 40 | 0.8 | 0.50 |
| -0.260 | -0.233 | 0.585 | 0.415 | 0.831 | 41 | 0.8 | 0.50 |
| -0.233 | -0.207 | 0.576 | 0.424 | 0.849 | 42 | 0.8 | 0.50 |
| -0.207 | -0.181 | 0.567 | 0.433 | 0.867 | 43 | 0.8 | 0.50 |
| -0.181 | -0.155 | 0.558 | 0.442 | 0.884 | 44 | 0.8 | 0.50 |
| -0.155 | -0.129 | 0.549 | 0.451 | 0.902 | 45 | 0.8 | 0.50 |
| -0.129 | -0.103 | 0.540 | 0.460 | 0.920 | 46 | 0.8 | 0.50 |
| -0.103 | -0.077 | 0.531 | 0.469 | 0.938 | 47 | 0.8 | 0.50 |
| -0.077 | -0.051 | 0.522 | 0.478 | 0.955 | 48 | 0.8 | 0.50 |
| -0.051 | -0.026 | 0.513 | 0.487 | 0.973 | 49 | 0.8 | 0.50 |
| -0.026 | 0.000 | 0.504 | 0.496 | 0.991 | 50 | 0.8 | 0.50 |
| 0.000 | 0.026 | 0.496 | 0.504 | 1.009 | 51 | 0.8 | 0.50 |
| 0.026 | 0.051 | 0.487 | 0.513 | 1.027 | 52 | 0.8 | 0.50 |
| 0.051 | 0.077 | 0.478 | 0.522 | 1.045 | 53 | 0.8 | 0.50 |
| 0.077 | 0.103 | 0.469 | 0.531 | 1.062 | 54 | 0.8 | 0.50 |
| 0.103 | 0.129 | 0.460 | 0.540 | 1.080 | 55 | 0.8 | 0.50 |
| 0.129 | 0.155 | 0.451 | 0.549 | 1.098 | 56 | 0.8 | 0.50 |
| 0.155 | 0.181 | 0.442 | 0.558 | 1.116 | 57 | 0.8 | 0.50 |
| 0.181 | 0.207 | 0.433 | 0.567 | 1.133 | 58 | 0.8 | 0.50 |
| 0.207 | 0.233 | 0.424 | 0.576 | 1.151 | 59 | 0.8 | 0.50 |
| 0.233 | 0.260 | 0.415 | 0.585 | 1.169 | 60 | 0.8 | 0.50 |
| 0.260 | 0.286 | 0.407 | 0.593 | 1.187 | 61 | 0.8 | 0.50 |
| 0.286 | 0.313 | 0.398 | 0.602 | 1.205 | 62 | 0.8 | 0.50 |
| 0.313 | 0.340 | 0.389 | 0.611 | 1.222 | 63 | 0.8 | 0.50 |
| 0.340 | 0.367 | 0.380 | 0.620 | 1.240 | 64 | 0.8 | 0.50 |
| 0.367 | 0.395 | 0.371 | 0.629 | 1.258 | 65 | 0.8 | 0.50 |
| 0.395 | 0.422 | 0.362 | 0.638 | 1.275 | 66 | 0.8 | 0.50 |
| 0.422 | 0.450 | 0.353 | 0.647 | 1.293 | 67 | 0.8 | 0.50 |
| 0.450 | 0.478 | 0.345 | 0.655 | 1.311 | 68 | 0.8 | 0.50 |
| 0.478 | 0.507 | 0.336 | 0.664 | 1.329 | 69 | 0.8 | 0.50 |
| 0.507 | 0.536 | 0.327 | 0.673 | 1.346 | 70 | 0.8 | 0.50 |
| 0.536 | 0.565 | 0.318 | 0.682 | 1.364 | 71 | 0.8 | 0.50 |
| 0.565 | 0.595 | 0.309 | 0.691 | 1.382 | 72 | 0.8 | 0.50 |
| 0.595 | 0.626 | 0.300 | 0.700 | 1.399 | 73 | 0.8 | 0.50 |
| 0.626 | 0.657 | 0.292 | 0.708 | 1.417 | 74 | 0.8 | 0.50 |
| 0.657 | 0.688 | 0.283 | 0.717 | 1.435 | 75 | 0.8 | 0.50 |
| 0.688 | 0.720 | 0.274 | 0.726 | 1.452 | 76 | 0.8 | 0.50 |
| 0.720 | 0.753 | 0.265 | 0.735 | 1.470 | 77 | 0.8 | 0.50 |
| 0.753 | 0.786 | 0.256 | 0.744 | 1.488 | 78 | 0.8 | 0.50 |
| 0.786 | 0.821 | 0.247 | 0.753 | 1.505 | 79 | 0.8 | 0.50 |
| 0.821 | 0.856 | 0.238 | 0.762 | 1.523 | 80 | 0.8 | 0.50 |
| 0.856 | 0.892 | 0.230 | 0.770 | 1.541 | 81 | 0.8 | 0.50 |
| 0.892 | 0.930 | 0.221 | 0.779 | 1.559 | 82 | 0.8 | 0.50 |
| 0.930 | 0.968 | 0.212 | 0.788 | 1.576 | 83 | 0.8 | 0.50 |
| 0.968 | 1.008 | 0.203 | 0.797 | 1.594 | 84 | 0.8 | 0.50 |
| 1.008 | 1.050 | 0.194 | 0.806 | 1.612 | 85 | 0.8 | 0.50 |
| 1.050 | 1.093 | 0.185 | 0.815 | 1.630 | 86 | 0.8 | 0.50 |
| 1.093 | 1.139 | 0.176 | 0.824 | 1.649 | 87 | 0.8 | 0.50 |
| 1.139 | 1.187 | 0.167 | 0.833 | 1.667 | 88 | 0.8 | 0.50 |
| 1.187 | 1.237 | 0.157 | 0.843 | 1.685 | 89 | 0.8 | 0.50 |
| 1.237 | 1.291 | 0.148 | 0.852 | 1.704 | 90 | 0.8 | 0.50 |
| 1.291 | 1.349 | 0.138 | 0.862 | 1.723 | 91 | 0.8 | 0.50 |
| 1.349 | 1.412 | 0.129 | 0.871 | 1.742 | 92 | 0.8 | 0.50 |
| 1.412 | 1.480 | 0.119 | 0.881 | 1.762 | 93 | 0.8 | 0.50 |
| 1.480 | 1.557 | 0.109 | 0.891 | 1.782 | 94 | 0.8 | 0.50 |
| 1.557 | 1.643 | 0.098 | 0.902 | 1.803 | 95 | 0.8 | 0.50 |
| 1.643 | 1.744 | 0.088 | 0.912 | 1.825 | 96 | 0.8 | 0.50 |
| 1.744 | 1.868 | 0.076 | 0.924 | 1.848 | 97 | 0.8 | 0.50 |
| 1.868 | 2.031 | 0.063 | 0.937 | 1.873 | 98 | 0.8 | 0.50 |
| 2.031 | 2.286 | 0.049 | 0.951 | 1.902 | 99 | 0.8 | 0.50 |
| 2.286 | Inf | 0.028 | 0.972 | 1.943 | 100 | 0.8 | 0.50 |
| -Inf | -2.194 | 0.997 | 0.003 | 0.006 | 1 | 0.9 | 0.50 |
| -2.194 | -1.970 | 0.994 | 0.006 | 0.013 | 2 | 0.9 | 0.50 |
| -1.970 | -1.826 | 0.990 | 0.010 | 0.019 | 3 | 0.9 | 0.50 |
| -1.826 | -1.716 | 0.987 | 0.013 | 0.026 | 4 | 0.9 | 0.50 |
| -1.716 | -1.625 | 0.983 | 0.017 | 0.033 | 5 | 0.9 | 0.50 |
| -1.625 | -1.547 | 0.980 | 0.020 | 0.041 | 6 | 0.9 | 0.50 |
| -1.547 | -1.478 | 0.976 | 0.024 | 0.048 | 7 | 0.9 | 0.50 |
| -1.478 | -1.416 | 0.972 | 0.028 | 0.057 | 8 | 0.9 | 0.50 |
| -1.416 | -1.359 | 0.967 | 0.033 | 0.065 | 9 | 0.9 | 0.50 |
| -1.359 | -1.305 | 0.963 | 0.037 | 0.074 | 10 | 0.9 | 0.50 |
| -1.305 | -1.255 | 0.958 | 0.042 | 0.084 | 11 | 0.9 | 0.50 |
| -1.255 | -1.208 | 0.953 | 0.047 | 0.094 | 12 | 0.9 | 0.50 |
| -1.208 | -1.163 | 0.948 | 0.052 | 0.104 | 13 | 0.9 | 0.50 |
| -1.163 | -1.121 | 0.942 | 0.058 | 0.115 | 14 | 0.9 | 0.50 |
| -1.121 | -1.080 | 0.936 | 0.064 | 0.127 | 15 | 0.9 | 0.50 |
| -1.080 | -1.040 | 0.930 | 0.070 | 0.139 | 16 | 0.9 | 0.50 |
| -1.040 | -1.002 | 0.924 | 0.076 | 0.152 | 17 | 0.9 | 0.50 |
| -1.002 | -0.964 | 0.917 | 0.083 | 0.166 | 18 | 0.9 | 0.50 |
| -0.964 | -0.928 | 0.910 | 0.090 | 0.180 | 19 | 0.9 | 0.50 |
| -0.928 | -0.893 | 0.903 | 0.097 | 0.195 | 20 | 0.9 | 0.50 |
| -0.893 | -0.858 | 0.895 | 0.105 | 0.210 | 21 | 0.9 | 0.50 |
| -0.858 | -0.824 | 0.887 | 0.113 | 0.227 | 22 | 0.9 | 0.50 |
| -0.824 | -0.791 | 0.878 | 0.122 | 0.244 | 23 | 0.9 | 0.50 |
| -0.791 | -0.759 | 0.869 | 0.131 | 0.261 | 24 | 0.9 | 0.50 |
| -0.759 | -0.727 | 0.860 | 0.140 | 0.280 | 25 | 0.9 | 0.50 |
| -0.727 | -0.695 | 0.850 | 0.150 | 0.299 | 26 | 0.9 | 0.50 |
| -0.695 | -0.664 | 0.840 | 0.160 | 0.319 | 27 | 0.9 | 0.50 |
| -0.664 | -0.633 | 0.830 | 0.170 | 0.340 | 28 | 0.9 | 0.50 |
| -0.633 | -0.602 | 0.819 | 0.181 | 0.362 | 29 | 0.9 | 0.50 |
| -0.602 | -0.572 | 0.808 | 0.192 | 0.384 | 30 | 0.9 | 0.50 |
| -0.572 | -0.542 | 0.796 | 0.204 | 0.408 | 31 | 0.9 | 0.50 |
| -0.542 | -0.512 | 0.784 | 0.216 | 0.432 | 32 | 0.9 | 0.50 |
| -0.512 | -0.483 | 0.772 | 0.228 | 0.457 | 33 | 0.9 | 0.50 |
| -0.483 | -0.454 | 0.759 | 0.241 | 0.483 | 34 | 0.9 | 0.50 |
| -0.454 | -0.425 | 0.745 | 0.255 | 0.509 | 35 | 0.9 | 0.50 |
| -0.425 | -0.396 | 0.732 | 0.268 | 0.537 | 36 | 0.9 | 0.50 |
| -0.396 | -0.367 | 0.718 | 0.282 | 0.565 | 37 | 0.9 | 0.50 |
| -0.367 | -0.338 | 0.703 | 0.297 | 0.594 | 38 | 0.9 | 0.50 |
| -0.338 | -0.310 | 0.688 | 0.312 | 0.623 | 39 | 0.9 | 0.50 |
| -0.310 | -0.281 | 0.673 | 0.327 | 0.654 | 40 | 0.9 | 0.50 |
| -0.281 | -0.253 | 0.658 | 0.342 | 0.684 | 41 | 0.9 | 0.50 |
| -0.253 | -0.225 | 0.642 | 0.358 | 0.716 | 42 | 0.9 | 0.50 |
| -0.225 | -0.196 | 0.626 | 0.374 | 0.748 | 43 | 0.9 | 0.50 |
| -0.196 | -0.168 | 0.610 | 0.390 | 0.781 | 44 | 0.9 | 0.50 |
| -0.168 | -0.140 | 0.593 | 0.407 | 0.814 | 45 | 0.9 | 0.50 |
| -0.140 | -0.112 | 0.577 | 0.423 | 0.847 | 46 | 0.9 | 0.50 |
| -0.112 | -0.084 | 0.560 | 0.440 | 0.881 | 47 | 0.9 | 0.50 |
| -0.084 | -0.056 | 0.543 | 0.457 | 0.915 | 48 | 0.9 | 0.50 |
| -0.056 | -0.028 | 0.526 | 0.474 | 0.949 | 49 | 0.9 | 0.50 |
| -0.028 | 0.000 | 0.509 | 0.491 | 0.983 | 50 | 0.9 | 0.50 |
| 0.000 | 0.028 | 0.491 | 0.509 | 1.017 | 51 | 0.9 | 0.50 |
| 0.028 | 0.056 | 0.474 | 0.526 | 1.051 | 52 | 0.9 | 0.50 |
| 0.056 | 0.084 | 0.457 | 0.543 | 1.085 | 53 | 0.9 | 0.50 |
| 0.084 | 0.112 | 0.440 | 0.560 | 1.119 | 54 | 0.9 | 0.50 |
| 0.112 | 0.140 | 0.423 | 0.577 | 1.153 | 55 | 0.9 | 0.50 |
| 0.140 | 0.168 | 0.407 | 0.593 | 1.186 | 56 | 0.9 | 0.50 |
| 0.168 | 0.196 | 0.390 | 0.610 | 1.219 | 57 | 0.9 | 0.50 |
| 0.196 | 0.225 | 0.374 | 0.626 | 1.252 | 58 | 0.9 | 0.50 |
| 0.225 | 0.253 | 0.358 | 0.642 | 1.284 | 59 | 0.9 | 0.50 |
| 0.253 | 0.281 | 0.342 | 0.658 | 1.316 | 60 | 0.9 | 0.50 |
| 0.281 | 0.310 | 0.327 | 0.673 | 1.346 | 61 | 0.9 | 0.50 |
| 0.310 | 0.338 | 0.312 | 0.688 | 1.377 | 62 | 0.9 | 0.50 |
| 0.338 | 0.367 | 0.297 | 0.703 | 1.406 | 63 | 0.9 | 0.50 |
| 0.367 | 0.396 | 0.282 | 0.718 | 1.435 | 64 | 0.9 | 0.50 |
| 0.396 | 0.425 | 0.268 | 0.732 | 1.463 | 65 | 0.9 | 0.50 |
| 0.425 | 0.454 | 0.255 | 0.745 | 1.491 | 66 | 0.9 | 0.50 |
| 0.454 | 0.483 | 0.241 | 0.759 | 1.517 | 67 | 0.9 | 0.50 |
| 0.483 | 0.512 | 0.228 | 0.772 | 1.543 | 68 | 0.9 | 0.50 |
| 0.512 | 0.542 | 0.216 | 0.784 | 1.568 | 69 | 0.9 | 0.50 |
| 0.542 | 0.572 | 0.204 | 0.796 | 1.592 | 70 | 0.9 | 0.50 |
| 0.572 | 0.602 | 0.192 | 0.808 | 1.616 | 71 | 0.9 | 0.50 |
| 0.602 | 0.633 | 0.181 | 0.819 | 1.638 | 72 | 0.9 | 0.50 |
| 0.633 | 0.664 | 0.170 | 0.830 | 1.660 | 73 | 0.9 | 0.50 |
| 0.664 | 0.695 | 0.160 | 0.840 | 1.681 | 74 | 0.9 | 0.50 |
| 0.695 | 0.727 | 0.150 | 0.850 | 1.701 | 75 | 0.9 | 0.50 |
| 0.727 | 0.759 | 0.140 | 0.860 | 1.720 | 76 | 0.9 | 0.50 |
| 0.759 | 0.791 | 0.131 | 0.869 | 1.739 | 77 | 0.9 | 0.50 |
| 0.791 | 0.824 | 0.122 | 0.878 | 1.756 | 78 | 0.9 | 0.50 |
| 0.824 | 0.858 | 0.113 | 0.887 | 1.773 | 79 | 0.9 | 0.50 |
| 0.858 | 0.893 | 0.105 | 0.895 | 1.790 | 80 | 0.9 | 0.50 |
| 0.893 | 0.928 | 0.097 | 0.903 | 1.805 | 81 | 0.9 | 0.50 |
| 0.928 | 0.964 | 0.090 | 0.910 | 1.820 | 82 | 0.9 | 0.50 |
| 0.964 | 1.002 | 0.083 | 0.917 | 1.834 | 83 | 0.9 | 0.50 |
| 1.002 | 1.040 | 0.076 | 0.924 | 1.848 | 84 | 0.9 | 0.50 |
| 1.040 | 1.080 | 0.070 | 0.930 | 1.861 | 85 | 0.9 | 0.50 |
| 1.080 | 1.121 | 0.064 | 0.936 | 1.873 | 86 | 0.9 | 0.50 |
| 1.121 | 1.163 | 0.058 | 0.942 | 1.885 | 87 | 0.9 | 0.50 |
| 1.163 | 1.208 | 0.052 | 0.948 | 1.896 | 88 | 0.9 | 0.50 |
| 1.208 | 1.255 | 0.047 | 0.953 | 1.906 | 89 | 0.9 | 0.50 |
| 1.255 | 1.305 | 0.042 | 0.958 | 1.916 | 90 | 0.9 | 0.50 |
| 1.305 | 1.359 | 0.037 | 0.963 | 1.926 | 91 | 0.9 | 0.50 |
| 1.359 | 1.416 | 0.033 | 0.967 | 1.935 | 92 | 0.9 | 0.50 |
| 1.416 | 1.478 | 0.028 | 0.972 | 1.943 | 93 | 0.9 | 0.50 |
| 1.478 | 1.547 | 0.024 | 0.976 | 1.952 | 94 | 0.9 | 0.50 |
| 1.547 | 1.625 | 0.020 | 0.980 | 1.959 | 95 | 0.9 | 0.50 |
| 1.625 | 1.716 | 0.017 | 0.983 | 1.967 | 96 | 0.9 | 0.50 |
| 1.716 | 1.826 | 0.013 | 0.987 | 1.974 | 97 | 0.9 | 0.50 |
| 1.826 | 1.970 | 0.010 | 0.990 | 1.981 | 98 | 0.9 | 0.50 |
| 1.970 | 2.194 | 0.006 | 0.994 | 1.987 | 99 | 0.9 | 0.50 |
| 2.194 | Inf | 0.003 | 0.997 | 1.994 | 100 | 0.9 | 0.50 |
1.3.2 Continuous outcomes
Show code
# Thank you for Alex Gillet for her work developing this code.
mean_sd_quant.f <- function(PRS_R2=0.641, Outcome_mean=1, Outcome_sd=1, n_quantile=20){
### PRS quantiles with a continuous phenotype (Y)
library(tmvtnorm)
###
E_PRS = 0
SD_PRS = sqrt(1)
E_phenotype = Outcome_mean
SD_phenotype = Outcome_sd
by_quant<-1/(n_quantile)
PRS_quantile_bounds <- qnorm(p=seq(0, 1, by=by_quant), mean= E_PRS, sd= SD_PRS)
lower_PRS_vec <- PRS_quantile_bounds[1:n_quantile]
upper_PRS_vec <- PRS_quantile_bounds[2:(n_quantile+1)]
mean_vec <- c(E_phenotype, E_PRS)
sigma_mat <- matrix(sqrt(PRS_R2)*SD_phenotype*SD_PRS, nrow=2, ncol=2)
sigma_mat[1,1] <- SD_phenotype^2
sigma_mat[2,2] <- SD_PRS^2
### mean of phenotype within the truncated PRS distribution
out_mean_Y <- rep(0, n_quantile)
### SD of phenotype within the truncated PRS distribution
out_SD_Y <- rep(0, n_quantile)
### cov of Y and PRS given truncation on PRS
out_cov_Y_PRS <- rep(0, n_quantile)
### SD of PRS given truncation on PRS
out_SD_PRS <- rep(0, n_quantile)
### mean PRS given truncation on PRS
out_mean_PRS <- rep(0, n_quantile)
for(i in 1:n_quantile){
distribution_i <- mtmvnorm(mean = mean_vec,
sigma = sigma_mat,
lower = c(-Inf, lower_PRS_vec[i]),
upper = c(Inf, upper_PRS_vec[i]),
doComputeVariance=TRUE,
pmvnorm.algorithm=GenzBretz())
out_mean_Y[i] <- distribution_i$tmean[1]
out_mean_PRS[i] <- distribution_i$tmean[2]
out_SD_Y[i] <- sqrt(distribution_i$tvar[1,1])
out_SD_PRS[i] <- sqrt(distribution_i$tvar[2,2])
out_cov_Y_PRS[i] <- distribution_i$tvar[1,2]
}
out<-data.frame(q=1:n_quantile,
q_min=lower_PRS_vec,
q_max=upper_PRS_vec,
x_mean=out_mean_Y,
x_sd=out_SD_Y)
return(out)
out_mean_Y
out_SD_Y
out_mean_PRS
out_SD_PRS
out_cov_Y_PRS
}
r2<-as.character(seq(0.1, 0.9, by=0.2))
library(ggplot2)
library(cowplot)
plot_list<-list()
res_all<-NULL
res_j<-NULL
for(j in r2){
res_j<-mean_sd_quant.f(PRS_R2=as.numeric(j), Outcome_mean=0, Outcome_sd=1, n_quantile=20)
res_j$Quantile<-1:nrow(res_j)
res_j$r2<-j
res_j$mean<-0
res_j$sd<-i
res_all<-rbind(res_all, res_j)
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Continuous_sim.png'), units='px', res=300, width=2000, height=1000)
ggplot(res_all, aes(x=Quantile, y=x_mean, colour=r2)) +
geom_point(position=position_dodge(.5)) +
geom_errorbar(aes(ymin=x_mean-x_sd, ymax=x_mean+x_sd), width=.2, position=position_dodge(.5), alpha=0.8) +
labs(y="Mean (SD)", colour=expression(paste(italic("R")^2))) +
theme_half_open() +
background_grid()
dev.off()
write.csv(res_all, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Continuous_sim.csv', row.names=F, quote=F)Show results

| q_min | q_max | p_control | p_case | OR | Quantile | auc | k |
|---|---|---|---|---|---|---|---|
| -Inf | -2.326 | 0.996 | 0.004 | 0.366 | 1 | 0.6 | 0.01 |
| -2.326 | -2.054 | 0.996 | 0.004 | 0.433 | 2 | 0.6 | 0.01 |
| -2.054 | -1.881 | 0.995 | 0.005 | 0.467 | 3 | 0.6 | 0.01 |
| -1.881 | -1.751 | 0.995 | 0.005 | 0.493 | 4 | 0.6 | 0.01 |
| -1.751 | -1.645 | 0.995 | 0.005 | 0.514 | 5 | 0.6 | 0.01 |
| -1.645 | -1.555 | 0.995 | 0.005 | 0.532 | 6 | 0.6 | 0.01 |
| -1.555 | -1.476 | 0.995 | 0.005 | 0.548 | 7 | 0.6 | 0.01 |
| -1.476 | -1.405 | 0.994 | 0.006 | 0.563 | 8 | 0.6 | 0.01 |
| -1.405 | -1.341 | 0.994 | 0.006 | 0.577 | 9 | 0.6 | 0.01 |
| -1.341 | -1.281 | 0.994 | 0.006 | 0.589 | 10 | 0.6 | 0.01 |
| -1.281 | -1.226 | 0.994 | 0.006 | 0.602 | 11 | 0.6 | 0.01 |
| -1.226 | -1.175 | 0.994 | 0.006 | 0.613 | 12 | 0.6 | 0.01 |
| -1.175 | -1.126 | 0.994 | 0.006 | 0.624 | 13 | 0.6 | 0.01 |
| -1.126 | -1.080 | 0.994 | 0.006 | 0.635 | 14 | 0.6 | 0.01 |
| -1.080 | -1.036 | 0.994 | 0.006 | 0.645 | 15 | 0.6 | 0.01 |
| -1.036 | -0.994 | 0.993 | 0.007 | 0.655 | 16 | 0.6 | 0.01 |
| -0.994 | -0.954 | 0.993 | 0.007 | 0.664 | 17 | 0.6 | 0.01 |
| -0.954 | -0.915 | 0.993 | 0.007 | 0.674 | 18 | 0.6 | 0.01 |
| -0.915 | -0.878 | 0.993 | 0.007 | 0.683 | 19 | 0.6 | 0.01 |
| -0.878 | -0.842 | 0.993 | 0.007 | 0.692 | 20 | 0.6 | 0.01 |
| -0.842 | -0.806 | 0.993 | 0.007 | 0.701 | 21 | 0.6 | 0.01 |
| -0.806 | -0.772 | 0.993 | 0.007 | 0.710 | 22 | 0.6 | 0.01 |
| -0.772 | -0.739 | 0.993 | 0.007 | 0.718 | 23 | 0.6 | 0.01 |
| -0.739 | -0.706 | 0.993 | 0.007 | 0.727 | 24 | 0.6 | 0.01 |
| -0.706 | -0.675 | 0.993 | 0.007 | 0.735 | 25 | 0.6 | 0.01 |
| -0.675 | -0.643 | 0.993 | 0.007 | 0.743 | 26 | 0.6 | 0.01 |
| -0.643 | -0.613 | 0.992 | 0.008 | 0.752 | 27 | 0.6 | 0.01 |
| -0.613 | -0.583 | 0.992 | 0.008 | 0.760 | 28 | 0.6 | 0.01 |
| -0.583 | -0.553 | 0.992 | 0.008 | 0.768 | 29 | 0.6 | 0.01 |
| -0.553 | -0.524 | 0.992 | 0.008 | 0.776 | 30 | 0.6 | 0.01 |
| -0.524 | -0.496 | 0.992 | 0.008 | 0.784 | 31 | 0.6 | 0.01 |
| -0.496 | -0.468 | 0.992 | 0.008 | 0.792 | 32 | 0.6 | 0.01 |
| -0.468 | -0.440 | 0.992 | 0.008 | 0.800 | 33 | 0.6 | 0.01 |
| -0.440 | -0.413 | 0.992 | 0.008 | 0.808 | 34 | 0.6 | 0.01 |
| -0.413 | -0.385 | 0.992 | 0.008 | 0.815 | 35 | 0.6 | 0.01 |
| -0.385 | -0.359 | 0.992 | 0.008 | 0.823 | 36 | 0.6 | 0.01 |
| -0.359 | -0.332 | 0.992 | 0.008 | 0.831 | 37 | 0.6 | 0.01 |
| -0.332 | -0.306 | 0.992 | 0.008 | 0.839 | 38 | 0.6 | 0.01 |
| -0.306 | -0.279 | 0.992 | 0.008 | 0.847 | 39 | 0.6 | 0.01 |
| -0.279 | -0.253 | 0.991 | 0.009 | 0.855 | 40 | 0.6 | 0.01 |
| -0.253 | -0.228 | 0.991 | 0.009 | 0.863 | 41 | 0.6 | 0.01 |
| -0.228 | -0.202 | 0.991 | 0.009 | 0.871 | 42 | 0.6 | 0.01 |
| -0.202 | -0.176 | 0.991 | 0.009 | 0.879 | 43 | 0.6 | 0.01 |
| -0.176 | -0.151 | 0.991 | 0.009 | 0.887 | 44 | 0.6 | 0.01 |
| -0.151 | -0.126 | 0.991 | 0.009 | 0.895 | 45 | 0.6 | 0.01 |
| -0.126 | -0.101 | 0.991 | 0.009 | 0.903 | 46 | 0.6 | 0.01 |
| -0.101 | -0.075 | 0.991 | 0.009 | 0.911 | 47 | 0.6 | 0.01 |
| -0.075 | -0.050 | 0.991 | 0.009 | 0.919 | 48 | 0.6 | 0.01 |
| -0.050 | -0.025 | 0.991 | 0.009 | 0.927 | 49 | 0.6 | 0.01 |
| -0.025 | 0.000 | 0.991 | 0.009 | 0.935 | 50 | 0.6 | 0.01 |
| 0.000 | 0.025 | 0.991 | 0.009 | 0.944 | 51 | 0.6 | 0.01 |
| 0.025 | 0.050 | 0.990 | 0.010 | 0.952 | 52 | 0.6 | 0.01 |
| 0.050 | 0.075 | 0.990 | 0.010 | 0.961 | 53 | 0.6 | 0.01 |
| 0.075 | 0.100 | 0.990 | 0.010 | 0.969 | 54 | 0.6 | 0.01 |
| 0.100 | 0.126 | 0.990 | 0.010 | 0.978 | 55 | 0.6 | 0.01 |
| 0.126 | 0.151 | 0.990 | 0.010 | 0.987 | 56 | 0.6 | 0.01 |
| 0.151 | 0.176 | 0.990 | 0.010 | 0.996 | 57 | 0.6 | 0.01 |
| 0.176 | 0.202 | 0.990 | 0.010 | 1.005 | 58 | 0.6 | 0.01 |
| 0.202 | 0.227 | 0.990 | 0.010 | 1.014 | 59 | 0.6 | 0.01 |
| 0.227 | 0.253 | 0.990 | 0.010 | 1.023 | 60 | 0.6 | 0.01 |
| 0.253 | 0.279 | 0.990 | 0.010 | 1.033 | 61 | 0.6 | 0.01 |
| 0.279 | 0.305 | 0.990 | 0.010 | 1.042 | 62 | 0.6 | 0.01 |
| 0.305 | 0.332 | 0.989 | 0.011 | 1.052 | 63 | 0.6 | 0.01 |
| 0.332 | 0.358 | 0.989 | 0.011 | 1.062 | 64 | 0.6 | 0.01 |
| 0.358 | 0.385 | 0.989 | 0.011 | 1.072 | 65 | 0.6 | 0.01 |
| 0.385 | 0.412 | 0.989 | 0.011 | 1.082 | 66 | 0.6 | 0.01 |
| 0.412 | 0.440 | 0.989 | 0.011 | 1.093 | 67 | 0.6 | 0.01 |
| 0.440 | 0.468 | 0.989 | 0.011 | 1.104 | 68 | 0.6 | 0.01 |
| 0.468 | 0.496 | 0.989 | 0.011 | 1.115 | 69 | 0.6 | 0.01 |
| 0.496 | 0.524 | 0.989 | 0.011 | 1.126 | 70 | 0.6 | 0.01 |
| 0.524 | 0.553 | 0.989 | 0.011 | 1.138 | 71 | 0.6 | 0.01 |
| 0.553 | 0.583 | 0.989 | 0.011 | 1.149 | 72 | 0.6 | 0.01 |
| 0.583 | 0.613 | 0.988 | 0.012 | 1.162 | 73 | 0.6 | 0.01 |
| 0.613 | 0.643 | 0.988 | 0.012 | 1.174 | 74 | 0.6 | 0.01 |
| 0.643 | 0.674 | 0.988 | 0.012 | 1.187 | 75 | 0.6 | 0.01 |
| 0.674 | 0.706 | 0.988 | 0.012 | 1.200 | 76 | 0.6 | 0.01 |
| 0.706 | 0.739 | 0.988 | 0.012 | 1.214 | 77 | 0.6 | 0.01 |
| 0.739 | 0.772 | 0.988 | 0.012 | 1.228 | 78 | 0.6 | 0.01 |
| 0.772 | 0.806 | 0.988 | 0.012 | 1.243 | 79 | 0.6 | 0.01 |
| 0.806 | 0.842 | 0.987 | 0.013 | 1.258 | 80 | 0.6 | 0.01 |
| 0.842 | 0.878 | 0.987 | 0.013 | 1.274 | 81 | 0.6 | 0.01 |
| 0.878 | 0.915 | 0.987 | 0.013 | 1.291 | 82 | 0.6 | 0.01 |
| 0.915 | 0.954 | 0.987 | 0.013 | 1.309 | 83 | 0.6 | 0.01 |
| 0.954 | 0.994 | 0.987 | 0.013 | 1.327 | 84 | 0.6 | 0.01 |
| 0.994 | 1.036 | 0.987 | 0.013 | 1.347 | 85 | 0.6 | 0.01 |
| 1.036 | 1.080 | 0.986 | 0.014 | 1.367 | 86 | 0.6 | 0.01 |
| 1.080 | 1.126 | 0.986 | 0.014 | 1.389 | 87 | 0.6 | 0.01 |
| 1.126 | 1.175 | 0.986 | 0.014 | 1.413 | 88 | 0.6 | 0.01 |
| 1.175 | 1.227 | 0.986 | 0.014 | 1.438 | 89 | 0.6 | 0.01 |
| 1.227 | 1.282 | 0.985 | 0.015 | 1.465 | 90 | 0.6 | 0.01 |
| 1.282 | 1.341 | 0.985 | 0.015 | 1.495 | 91 | 0.6 | 0.01 |
| 1.341 | 1.405 | 0.985 | 0.015 | 1.528 | 92 | 0.6 | 0.01 |
| 1.405 | 1.476 | 0.984 | 0.016 | 1.565 | 93 | 0.6 | 0.01 |
| 1.476 | 1.555 | 0.984 | 0.016 | 1.606 | 94 | 0.6 | 0.01 |
| 1.555 | 1.645 | 0.983 | 0.017 | 1.655 | 95 | 0.6 | 0.01 |
| 1.645 | 1.751 | 0.983 | 0.017 | 1.713 | 96 | 0.6 | 0.01 |
| 1.751 | 1.881 | 0.982 | 0.018 | 1.785 | 97 | 0.6 | 0.01 |
| 1.881 | 2.054 | 0.981 | 0.019 | 1.881 | 98 | 0.6 | 0.01 |
| 2.054 | 2.327 | 0.980 | 0.020 | 2.029 | 99 | 0.6 | 0.01 |
| 2.327 | Inf | 0.976 | 0.024 | 2.422 | 100 | 0.6 | 0.01 |
| -Inf | -2.324 | 0.999 | 0.001 | 0.109 | 1 | 0.7 | 0.01 |
| -2.324 | -2.052 | 0.998 | 0.002 | 0.153 | 2 | 0.7 | 0.01 |
| -2.052 | -1.879 | 0.998 | 0.002 | 0.179 | 3 | 0.7 | 0.01 |
| -1.879 | -1.749 | 0.998 | 0.002 | 0.200 | 4 | 0.7 | 0.01 |
| -1.749 | -1.644 | 0.998 | 0.002 | 0.218 | 5 | 0.7 | 0.01 |
| -1.644 | -1.554 | 0.998 | 0.002 | 0.235 | 6 | 0.7 | 0.01 |
| -1.554 | -1.475 | 0.998 | 0.002 | 0.250 | 7 | 0.7 | 0.01 |
| -1.475 | -1.404 | 0.997 | 0.003 | 0.264 | 8 | 0.7 | 0.01 |
| -1.404 | -1.340 | 0.997 | 0.003 | 0.277 | 9 | 0.7 | 0.01 |
| -1.340 | -1.281 | 0.997 | 0.003 | 0.290 | 10 | 0.7 | 0.01 |
| -1.281 | -1.226 | 0.997 | 0.003 | 0.303 | 11 | 0.7 | 0.01 |
| -1.226 | -1.175 | 0.997 | 0.003 | 0.315 | 12 | 0.7 | 0.01 |
| -1.175 | -1.126 | 0.997 | 0.003 | 0.327 | 13 | 0.7 | 0.01 |
| -1.126 | -1.080 | 0.997 | 0.003 | 0.339 | 14 | 0.7 | 0.01 |
| -1.080 | -1.036 | 0.996 | 0.004 | 0.350 | 15 | 0.7 | 0.01 |
| -1.036 | -0.994 | 0.996 | 0.004 | 0.361 | 16 | 0.7 | 0.01 |
| -0.994 | -0.954 | 0.996 | 0.004 | 0.373 | 17 | 0.7 | 0.01 |
| -0.954 | -0.915 | 0.996 | 0.004 | 0.384 | 18 | 0.7 | 0.01 |
| -0.915 | -0.878 | 0.996 | 0.004 | 0.395 | 19 | 0.7 | 0.01 |
| -0.878 | -0.842 | 0.996 | 0.004 | 0.405 | 20 | 0.7 | 0.01 |
| -0.842 | -0.806 | 0.996 | 0.004 | 0.416 | 21 | 0.7 | 0.01 |
| -0.806 | -0.772 | 0.996 | 0.004 | 0.427 | 22 | 0.7 | 0.01 |
| -0.772 | -0.739 | 0.996 | 0.004 | 0.438 | 23 | 0.7 | 0.01 |
| -0.739 | -0.706 | 0.996 | 0.004 | 0.449 | 24 | 0.7 | 0.01 |
| -0.706 | -0.675 | 0.995 | 0.005 | 0.460 | 25 | 0.7 | 0.01 |
| -0.675 | -0.644 | 0.995 | 0.005 | 0.470 | 26 | 0.7 | 0.01 |
| -0.644 | -0.613 | 0.995 | 0.005 | 0.481 | 27 | 0.7 | 0.01 |
| -0.613 | -0.583 | 0.995 | 0.005 | 0.492 | 28 | 0.7 | 0.01 |
| -0.583 | -0.554 | 0.995 | 0.005 | 0.503 | 29 | 0.7 | 0.01 |
| -0.554 | -0.525 | 0.995 | 0.005 | 0.514 | 30 | 0.7 | 0.01 |
| -0.525 | -0.496 | 0.995 | 0.005 | 0.525 | 31 | 0.7 | 0.01 |
| -0.496 | -0.468 | 0.995 | 0.005 | 0.536 | 32 | 0.7 | 0.01 |
| -0.468 | -0.440 | 0.995 | 0.005 | 0.547 | 33 | 0.7 | 0.01 |
| -0.440 | -0.413 | 0.994 | 0.006 | 0.559 | 34 | 0.7 | 0.01 |
| -0.413 | -0.386 | 0.994 | 0.006 | 0.570 | 35 | 0.7 | 0.01 |
| -0.386 | -0.359 | 0.994 | 0.006 | 0.582 | 36 | 0.7 | 0.01 |
| -0.359 | -0.332 | 0.994 | 0.006 | 0.593 | 37 | 0.7 | 0.01 |
| -0.332 | -0.306 | 0.994 | 0.006 | 0.605 | 38 | 0.7 | 0.01 |
| -0.306 | -0.280 | 0.994 | 0.006 | 0.617 | 39 | 0.7 | 0.01 |
| -0.280 | -0.254 | 0.994 | 0.006 | 0.629 | 40 | 0.7 | 0.01 |
| -0.254 | -0.228 | 0.994 | 0.006 | 0.641 | 41 | 0.7 | 0.01 |
| -0.228 | -0.202 | 0.993 | 0.007 | 0.653 | 42 | 0.7 | 0.01 |
| -0.202 | -0.177 | 0.993 | 0.007 | 0.666 | 43 | 0.7 | 0.01 |
| -0.177 | -0.152 | 0.993 | 0.007 | 0.678 | 44 | 0.7 | 0.01 |
| -0.152 | -0.126 | 0.993 | 0.007 | 0.691 | 45 | 0.7 | 0.01 |
| -0.126 | -0.101 | 0.993 | 0.007 | 0.704 | 46 | 0.7 | 0.01 |
| -0.101 | -0.076 | 0.993 | 0.007 | 0.717 | 47 | 0.7 | 0.01 |
| -0.076 | -0.051 | 0.993 | 0.007 | 0.731 | 48 | 0.7 | 0.01 |
| -0.051 | -0.026 | 0.993 | 0.007 | 0.744 | 49 | 0.7 | 0.01 |
| -0.026 | -0.001 | 0.992 | 0.008 | 0.758 | 50 | 0.7 | 0.01 |
| -0.001 | 0.024 | 0.992 | 0.008 | 0.772 | 51 | 0.7 | 0.01 |
| 0.024 | 0.050 | 0.992 | 0.008 | 0.787 | 52 | 0.7 | 0.01 |
| 0.050 | 0.075 | 0.992 | 0.008 | 0.801 | 53 | 0.7 | 0.01 |
| 0.075 | 0.100 | 0.992 | 0.008 | 0.816 | 54 | 0.7 | 0.01 |
| 0.100 | 0.125 | 0.992 | 0.008 | 0.832 | 55 | 0.7 | 0.01 |
| 0.125 | 0.150 | 0.992 | 0.008 | 0.847 | 56 | 0.7 | 0.01 |
| 0.150 | 0.176 | 0.991 | 0.009 | 0.863 | 57 | 0.7 | 0.01 |
| 0.176 | 0.201 | 0.991 | 0.009 | 0.880 | 58 | 0.7 | 0.01 |
| 0.201 | 0.227 | 0.991 | 0.009 | 0.896 | 59 | 0.7 | 0.01 |
| 0.227 | 0.253 | 0.991 | 0.009 | 0.914 | 60 | 0.7 | 0.01 |
| 0.253 | 0.279 | 0.991 | 0.009 | 0.931 | 61 | 0.7 | 0.01 |
| 0.279 | 0.305 | 0.991 | 0.009 | 0.949 | 62 | 0.7 | 0.01 |
| 0.305 | 0.331 | 0.990 | 0.010 | 0.968 | 63 | 0.7 | 0.01 |
| 0.331 | 0.358 | 0.990 | 0.010 | 0.987 | 64 | 0.7 | 0.01 |
| 0.358 | 0.385 | 0.990 | 0.010 | 1.007 | 65 | 0.7 | 0.01 |
| 0.385 | 0.412 | 0.990 | 0.010 | 1.027 | 66 | 0.7 | 0.01 |
| 0.412 | 0.439 | 0.990 | 0.010 | 1.048 | 67 | 0.7 | 0.01 |
| 0.439 | 0.467 | 0.989 | 0.011 | 1.069 | 68 | 0.7 | 0.01 |
| 0.467 | 0.495 | 0.989 | 0.011 | 1.091 | 69 | 0.7 | 0.01 |
| 0.495 | 0.524 | 0.989 | 0.011 | 1.114 | 70 | 0.7 | 0.01 |
| 0.524 | 0.553 | 0.989 | 0.011 | 1.138 | 71 | 0.7 | 0.01 |
| 0.553 | 0.582 | 0.988 | 0.012 | 1.163 | 72 | 0.7 | 0.01 |
| 0.582 | 0.612 | 0.988 | 0.012 | 1.189 | 73 | 0.7 | 0.01 |
| 0.612 | 0.643 | 0.988 | 0.012 | 1.215 | 74 | 0.7 | 0.01 |
| 0.643 | 0.674 | 0.988 | 0.012 | 1.243 | 75 | 0.7 | 0.01 |
| 0.674 | 0.706 | 0.987 | 0.013 | 1.272 | 76 | 0.7 | 0.01 |
| 0.706 | 0.738 | 0.987 | 0.013 | 1.303 | 77 | 0.7 | 0.01 |
| 0.738 | 0.772 | 0.987 | 0.013 | 1.335 | 78 | 0.7 | 0.01 |
| 0.772 | 0.806 | 0.986 | 0.014 | 1.368 | 79 | 0.7 | 0.01 |
| 0.806 | 0.841 | 0.986 | 0.014 | 1.403 | 80 | 0.7 | 0.01 |
| 0.841 | 0.878 | 0.986 | 0.014 | 1.441 | 81 | 0.7 | 0.01 |
| 0.878 | 0.915 | 0.985 | 0.015 | 1.480 | 82 | 0.7 | 0.01 |
| 0.915 | 0.954 | 0.985 | 0.015 | 1.522 | 83 | 0.7 | 0.01 |
| 0.954 | 0.994 | 0.984 | 0.016 | 1.567 | 84 | 0.7 | 0.01 |
| 0.994 | 1.036 | 0.984 | 0.016 | 1.615 | 85 | 0.7 | 0.01 |
| 1.036 | 1.080 | 0.983 | 0.017 | 1.666 | 86 | 0.7 | 0.01 |
| 1.080 | 1.126 | 0.983 | 0.017 | 1.722 | 87 | 0.7 | 0.01 |
| 1.126 | 1.175 | 0.982 | 0.018 | 1.783 | 88 | 0.7 | 0.01 |
| 1.175 | 1.227 | 0.982 | 0.018 | 1.849 | 89 | 0.7 | 0.01 |
| 1.227 | 1.282 | 0.981 | 0.019 | 1.922 | 90 | 0.7 | 0.01 |
| 1.282 | 1.341 | 0.980 | 0.020 | 2.004 | 91 | 0.7 | 0.01 |
| 1.341 | 1.405 | 0.979 | 0.021 | 2.097 | 92 | 0.7 | 0.01 |
| 1.405 | 1.476 | 0.978 | 0.022 | 2.202 | 93 | 0.7 | 0.01 |
| 1.476 | 1.556 | 0.977 | 0.023 | 2.325 | 94 | 0.7 | 0.01 |
| 1.556 | 1.646 | 0.975 | 0.025 | 2.472 | 95 | 0.7 | 0.01 |
| 1.646 | 1.752 | 0.973 | 0.027 | 2.654 | 96 | 0.7 | 0.01 |
| 1.752 | 1.882 | 0.971 | 0.029 | 2.889 | 97 | 0.7 | 0.01 |
| 1.882 | 2.056 | 0.968 | 0.032 | 3.219 | 98 | 0.7 | 0.01 |
| 2.056 | 2.330 | 0.962 | 0.038 | 3.760 | 99 | 0.7 | 0.01 |
| 2.330 | Inf | 0.945 | 0.055 | 5.458 | 100 | 0.7 | 0.01 |
| -Inf | -2.318 | 1.000 | 0.000 | 0.022 | 1 | 0.8 | 0.01 |
| -2.318 | -2.047 | 1.000 | 0.000 | 0.038 | 2 | 0.8 | 0.01 |
| -2.047 | -1.875 | 1.000 | 0.000 | 0.048 | 3 | 0.8 | 0.01 |
| -1.875 | -1.746 | 0.999 | 0.001 | 0.058 | 4 | 0.8 | 0.01 |
| -1.746 | -1.641 | 0.999 | 0.001 | 0.066 | 5 | 0.8 | 0.01 |
| -1.641 | -1.551 | 0.999 | 0.001 | 0.075 | 6 | 0.8 | 0.01 |
| -1.551 | -1.472 | 0.999 | 0.001 | 0.082 | 7 | 0.8 | 0.01 |
| -1.472 | -1.402 | 0.999 | 0.001 | 0.090 | 8 | 0.8 | 0.01 |
| -1.402 | -1.338 | 0.999 | 0.001 | 0.098 | 9 | 0.8 | 0.01 |
| -1.338 | -1.279 | 0.999 | 0.001 | 0.105 | 10 | 0.8 | 0.01 |
| -1.279 | -1.224 | 0.999 | 0.001 | 0.112 | 11 | 0.8 | 0.01 |
| -1.224 | -1.173 | 0.999 | 0.001 | 0.120 | 12 | 0.8 | 0.01 |
| -1.173 | -1.125 | 0.999 | 0.001 | 0.127 | 13 | 0.8 | 0.01 |
| -1.125 | -1.079 | 0.999 | 0.001 | 0.135 | 14 | 0.8 | 0.01 |
| -1.079 | -1.035 | 0.999 | 0.001 | 0.142 | 15 | 0.8 | 0.01 |
| -1.035 | -0.993 | 0.999 | 0.001 | 0.149 | 16 | 0.8 | 0.01 |
| -0.993 | -0.953 | 0.998 | 0.002 | 0.157 | 17 | 0.8 | 0.01 |
| -0.953 | -0.915 | 0.998 | 0.002 | 0.164 | 18 | 0.8 | 0.01 |
| -0.915 | -0.877 | 0.998 | 0.002 | 0.172 | 19 | 0.8 | 0.01 |
| -0.877 | -0.841 | 0.998 | 0.002 | 0.180 | 20 | 0.8 | 0.01 |
| -0.841 | -0.806 | 0.998 | 0.002 | 0.188 | 21 | 0.8 | 0.01 |
| -0.806 | -0.772 | 0.998 | 0.002 | 0.196 | 22 | 0.8 | 0.01 |
| -0.772 | -0.739 | 0.998 | 0.002 | 0.204 | 23 | 0.8 | 0.01 |
| -0.739 | -0.706 | 0.998 | 0.002 | 0.212 | 24 | 0.8 | 0.01 |
| -0.706 | -0.675 | 0.998 | 0.002 | 0.220 | 25 | 0.8 | 0.01 |
| -0.675 | -0.644 | 0.998 | 0.002 | 0.228 | 26 | 0.8 | 0.01 |
| -0.644 | -0.613 | 0.998 | 0.002 | 0.237 | 27 | 0.8 | 0.01 |
| -0.613 | -0.583 | 0.998 | 0.002 | 0.246 | 28 | 0.8 | 0.01 |
| -0.583 | -0.554 | 0.997 | 0.003 | 0.255 | 29 | 0.8 | 0.01 |
| -0.554 | -0.525 | 0.997 | 0.003 | 0.264 | 30 | 0.8 | 0.01 |
| -0.525 | -0.497 | 0.997 | 0.003 | 0.273 | 31 | 0.8 | 0.01 |
| -0.497 | -0.469 | 0.997 | 0.003 | 0.282 | 32 | 0.8 | 0.01 |
| -0.469 | -0.441 | 0.997 | 0.003 | 0.292 | 33 | 0.8 | 0.01 |
| -0.441 | -0.414 | 0.997 | 0.003 | 0.301 | 34 | 0.8 | 0.01 |
| -0.414 | -0.387 | 0.997 | 0.003 | 0.311 | 35 | 0.8 | 0.01 |
| -0.387 | -0.360 | 0.997 | 0.003 | 0.322 | 36 | 0.8 | 0.01 |
| -0.360 | -0.333 | 0.997 | 0.003 | 0.332 | 37 | 0.8 | 0.01 |
| -0.333 | -0.307 | 0.997 | 0.003 | 0.343 | 38 | 0.8 | 0.01 |
| -0.307 | -0.281 | 0.996 | 0.004 | 0.353 | 39 | 0.8 | 0.01 |
| -0.281 | -0.255 | 0.996 | 0.004 | 0.365 | 40 | 0.8 | 0.01 |
| -0.255 | -0.229 | 0.996 | 0.004 | 0.376 | 41 | 0.8 | 0.01 |
| -0.229 | -0.204 | 0.996 | 0.004 | 0.388 | 42 | 0.8 | 0.01 |
| -0.204 | -0.178 | 0.996 | 0.004 | 0.400 | 43 | 0.8 | 0.01 |
| -0.178 | -0.153 | 0.996 | 0.004 | 0.412 | 44 | 0.8 | 0.01 |
| -0.153 | -0.128 | 0.996 | 0.004 | 0.425 | 45 | 0.8 | 0.01 |
| -0.128 | -0.102 | 0.996 | 0.004 | 0.438 | 46 | 0.8 | 0.01 |
| -0.102 | -0.077 | 0.995 | 0.005 | 0.451 | 47 | 0.8 | 0.01 |
| -0.077 | -0.052 | 0.995 | 0.005 | 0.465 | 48 | 0.8 | 0.01 |
| -0.052 | -0.027 | 0.995 | 0.005 | 0.479 | 49 | 0.8 | 0.01 |
| -0.027 | -0.002 | 0.995 | 0.005 | 0.493 | 50 | 0.8 | 0.01 |
| -0.002 | 0.023 | 0.995 | 0.005 | 0.508 | 51 | 0.8 | 0.01 |
| 0.023 | 0.048 | 0.995 | 0.005 | 0.524 | 52 | 0.8 | 0.01 |
| 0.048 | 0.073 | 0.995 | 0.005 | 0.539 | 53 | 0.8 | 0.01 |
| 0.073 | 0.098 | 0.994 | 0.006 | 0.556 | 54 | 0.8 | 0.01 |
| 0.098 | 0.123 | 0.994 | 0.006 | 0.573 | 55 | 0.8 | 0.01 |
| 0.123 | 0.148 | 0.994 | 0.006 | 0.590 | 56 | 0.8 | 0.01 |
| 0.148 | 0.174 | 0.994 | 0.006 | 0.608 | 57 | 0.8 | 0.01 |
| 0.174 | 0.199 | 0.994 | 0.006 | 0.627 | 58 | 0.8 | 0.01 |
| 0.199 | 0.225 | 0.994 | 0.006 | 0.646 | 59 | 0.8 | 0.01 |
| 0.225 | 0.251 | 0.993 | 0.007 | 0.667 | 60 | 0.8 | 0.01 |
| 0.251 | 0.277 | 0.993 | 0.007 | 0.687 | 61 | 0.8 | 0.01 |
| 0.277 | 0.303 | 0.993 | 0.007 | 0.709 | 62 | 0.8 | 0.01 |
| 0.303 | 0.329 | 0.993 | 0.007 | 0.732 | 63 | 0.8 | 0.01 |
| 0.329 | 0.356 | 0.992 | 0.008 | 0.755 | 64 | 0.8 | 0.01 |
| 0.356 | 0.383 | 0.992 | 0.008 | 0.779 | 65 | 0.8 | 0.01 |
| 0.383 | 0.410 | 0.992 | 0.008 | 0.805 | 66 | 0.8 | 0.01 |
| 0.410 | 0.437 | 0.992 | 0.008 | 0.831 | 67 | 0.8 | 0.01 |
| 0.437 | 0.465 | 0.991 | 0.009 | 0.859 | 68 | 0.8 | 0.01 |
| 0.465 | 0.493 | 0.991 | 0.009 | 0.888 | 69 | 0.8 | 0.01 |
| 0.493 | 0.522 | 0.991 | 0.009 | 0.918 | 70 | 0.8 | 0.01 |
| 0.522 | 0.551 | 0.990 | 0.010 | 0.950 | 71 | 0.8 | 0.01 |
| 0.551 | 0.580 | 0.990 | 0.010 | 0.984 | 72 | 0.8 | 0.01 |
| 0.580 | 0.610 | 0.990 | 0.010 | 1.019 | 73 | 0.8 | 0.01 |
| 0.610 | 0.641 | 0.989 | 0.011 | 1.057 | 74 | 0.8 | 0.01 |
| 0.641 | 0.672 | 0.989 | 0.011 | 1.096 | 75 | 0.8 | 0.01 |
| 0.672 | 0.704 | 0.989 | 0.011 | 1.138 | 76 | 0.8 | 0.01 |
| 0.704 | 0.736 | 0.988 | 0.012 | 1.182 | 77 | 0.8 | 0.01 |
| 0.736 | 0.770 | 0.988 | 0.012 | 1.229 | 78 | 0.8 | 0.01 |
| 0.770 | 0.804 | 0.987 | 0.013 | 1.279 | 79 | 0.8 | 0.01 |
| 0.804 | 0.839 | 0.987 | 0.013 | 1.333 | 80 | 0.8 | 0.01 |
| 0.839 | 0.876 | 0.986 | 0.014 | 1.391 | 81 | 0.8 | 0.01 |
| 0.876 | 0.913 | 0.985 | 0.015 | 1.453 | 82 | 0.8 | 0.01 |
| 0.913 | 0.952 | 0.985 | 0.015 | 1.520 | 83 | 0.8 | 0.01 |
| 0.952 | 0.993 | 0.984 | 0.016 | 1.593 | 84 | 0.8 | 0.01 |
| 0.993 | 1.035 | 0.983 | 0.017 | 1.672 | 85 | 0.8 | 0.01 |
| 1.035 | 1.079 | 0.982 | 0.018 | 1.759 | 86 | 0.8 | 0.01 |
| 1.079 | 1.125 | 0.981 | 0.019 | 1.855 | 87 | 0.8 | 0.01 |
| 1.125 | 1.174 | 0.980 | 0.020 | 1.962 | 88 | 0.8 | 0.01 |
| 1.174 | 1.226 | 0.979 | 0.021 | 2.082 | 89 | 0.8 | 0.01 |
| 1.226 | 1.281 | 0.978 | 0.022 | 2.217 | 90 | 0.8 | 0.01 |
| 1.281 | 1.341 | 0.976 | 0.024 | 2.371 | 91 | 0.8 | 0.01 |
| 1.341 | 1.406 | 0.975 | 0.025 | 2.549 | 92 | 0.8 | 0.01 |
| 1.406 | 1.477 | 0.972 | 0.028 | 2.760 | 93 | 0.8 | 0.01 |
| 1.477 | 1.557 | 0.970 | 0.030 | 3.013 | 94 | 0.8 | 0.01 |
| 1.557 | 1.648 | 0.967 | 0.033 | 3.326 | 95 | 0.8 | 0.01 |
| 1.648 | 1.755 | 0.963 | 0.037 | 3.728 | 96 | 0.8 | 0.01 |
| 1.755 | 1.887 | 0.957 | 0.043 | 4.275 | 97 | 0.8 | 0.01 |
| 1.887 | 2.063 | 0.949 | 0.051 | 5.086 | 98 | 0.8 | 0.01 |
| 2.063 | 2.342 | 0.935 | 0.065 | 6.523 | 99 | 0.8 | 0.01 |
| 2.342 | Inf | 0.881 | 0.119 | 11.908 | 100 | 0.8 | 0.01 |
| -Inf | -2.304 | 1.000 | 0.000 | 0.002 | 1 | 0.9 | 0.01 |
| -2.304 | -2.035 | 1.000 | 0.000 | 0.004 | 2 | 0.9 | 0.01 |
| -2.035 | -1.864 | 1.000 | 0.000 | 0.006 | 3 | 0.9 | 0.01 |
| -1.864 | -1.736 | 1.000 | 0.000 | 0.007 | 4 | 0.9 | 0.01 |
| -1.736 | -1.632 | 1.000 | 0.000 | 0.009 | 5 | 0.9 | 0.01 |
| -1.632 | -1.543 | 1.000 | 0.000 | 0.011 | 6 | 0.9 | 0.01 |
| -1.543 | -1.465 | 1.000 | 0.000 | 0.013 | 7 | 0.9 | 0.01 |
| -1.465 | -1.395 | 1.000 | 0.000 | 0.015 | 8 | 0.9 | 0.01 |
| -1.395 | -1.332 | 1.000 | 0.000 | 0.016 | 9 | 0.9 | 0.01 |
| -1.332 | -1.273 | 1.000 | 0.000 | 0.018 | 10 | 0.9 | 0.01 |
| -1.273 | -1.219 | 1.000 | 0.000 | 0.020 | 11 | 0.9 | 0.01 |
| -1.219 | -1.168 | 1.000 | 0.000 | 0.022 | 12 | 0.9 | 0.01 |
| -1.168 | -1.120 | 1.000 | 0.000 | 0.025 | 13 | 0.9 | 0.01 |
| -1.120 | -1.075 | 1.000 | 0.000 | 0.027 | 14 | 0.9 | 0.01 |
| -1.075 | -1.032 | 1.000 | 0.000 | 0.029 | 15 | 0.9 | 0.01 |
| -1.032 | -0.990 | 1.000 | 0.000 | 0.031 | 16 | 0.9 | 0.01 |
| -0.990 | -0.950 | 1.000 | 0.000 | 0.034 | 17 | 0.9 | 0.01 |
| -0.950 | -0.912 | 1.000 | 0.000 | 0.036 | 18 | 0.9 | 0.01 |
| -0.912 | -0.875 | 1.000 | 0.000 | 0.039 | 19 | 0.9 | 0.01 |
| -0.875 | -0.839 | 1.000 | 0.000 | 0.042 | 20 | 0.9 | 0.01 |
| -0.839 | -0.804 | 1.000 | 0.000 | 0.044 | 21 | 0.9 | 0.01 |
| -0.804 | -0.771 | 1.000 | 0.000 | 0.047 | 22 | 0.9 | 0.01 |
| -0.771 | -0.738 | 0.999 | 0.001 | 0.050 | 23 | 0.9 | 0.01 |
| -0.738 | -0.705 | 0.999 | 0.001 | 0.053 | 24 | 0.9 | 0.01 |
| -0.705 | -0.674 | 0.999 | 0.001 | 0.057 | 25 | 0.9 | 0.01 |
| -0.674 | -0.643 | 0.999 | 0.001 | 0.060 | 26 | 0.9 | 0.01 |
| -0.643 | -0.613 | 0.999 | 0.001 | 0.064 | 27 | 0.9 | 0.01 |
| -0.613 | -0.583 | 0.999 | 0.001 | 0.067 | 28 | 0.9 | 0.01 |
| -0.583 | -0.554 | 0.999 | 0.001 | 0.071 | 29 | 0.9 | 0.01 |
| -0.554 | -0.526 | 0.999 | 0.001 | 0.075 | 30 | 0.9 | 0.01 |
| -0.526 | -0.497 | 0.999 | 0.001 | 0.079 | 31 | 0.9 | 0.01 |
| -0.497 | -0.470 | 0.999 | 0.001 | 0.083 | 32 | 0.9 | 0.01 |
| -0.470 | -0.442 | 0.999 | 0.001 | 0.087 | 33 | 0.9 | 0.01 |
| -0.442 | -0.415 | 0.999 | 0.001 | 0.092 | 34 | 0.9 | 0.01 |
| -0.415 | -0.388 | 0.999 | 0.001 | 0.096 | 35 | 0.9 | 0.01 |
| -0.388 | -0.361 | 0.999 | 0.001 | 0.101 | 36 | 0.9 | 0.01 |
| -0.361 | -0.335 | 0.999 | 0.001 | 0.106 | 37 | 0.9 | 0.01 |
| -0.335 | -0.309 | 0.999 | 0.001 | 0.112 | 38 | 0.9 | 0.01 |
| -0.309 | -0.283 | 0.999 | 0.001 | 0.117 | 39 | 0.9 | 0.01 |
| -0.283 | -0.257 | 0.999 | 0.001 | 0.123 | 40 | 0.9 | 0.01 |
| -0.257 | -0.232 | 0.999 | 0.001 | 0.129 | 41 | 0.9 | 0.01 |
| -0.232 | -0.206 | 0.999 | 0.001 | 0.135 | 42 | 0.9 | 0.01 |
| -0.206 | -0.181 | 0.999 | 0.001 | 0.141 | 43 | 0.9 | 0.01 |
| -0.181 | -0.156 | 0.999 | 0.001 | 0.148 | 44 | 0.9 | 0.01 |
| -0.156 | -0.131 | 0.998 | 0.002 | 0.155 | 45 | 0.9 | 0.01 |
| -0.131 | -0.106 | 0.998 | 0.002 | 0.162 | 46 | 0.9 | 0.01 |
| -0.106 | -0.081 | 0.998 | 0.002 | 0.170 | 47 | 0.9 | 0.01 |
| -0.081 | -0.056 | 0.998 | 0.002 | 0.178 | 48 | 0.9 | 0.01 |
| -0.056 | -0.031 | 0.998 | 0.002 | 0.186 | 49 | 0.9 | 0.01 |
| -0.031 | -0.006 | 0.998 | 0.002 | 0.195 | 50 | 0.9 | 0.01 |
| -0.006 | 0.019 | 0.998 | 0.002 | 0.204 | 51 | 0.9 | 0.01 |
| 0.019 | 0.043 | 0.998 | 0.002 | 0.213 | 52 | 0.9 | 0.01 |
| 0.043 | 0.068 | 0.998 | 0.002 | 0.223 | 53 | 0.9 | 0.01 |
| 0.068 | 0.093 | 0.998 | 0.002 | 0.234 | 54 | 0.9 | 0.01 |
| 0.093 | 0.118 | 0.998 | 0.002 | 0.245 | 55 | 0.9 | 0.01 |
| 0.118 | 0.144 | 0.997 | 0.003 | 0.256 | 56 | 0.9 | 0.01 |
| 0.144 | 0.169 | 0.997 | 0.003 | 0.269 | 57 | 0.9 | 0.01 |
| 0.169 | 0.194 | 0.997 | 0.003 | 0.281 | 58 | 0.9 | 0.01 |
| 0.194 | 0.220 | 0.997 | 0.003 | 0.295 | 59 | 0.9 | 0.01 |
| 0.220 | 0.245 | 0.997 | 0.003 | 0.309 | 60 | 0.9 | 0.01 |
| 0.245 | 0.271 | 0.997 | 0.003 | 0.324 | 61 | 0.9 | 0.01 |
| 0.271 | 0.297 | 0.997 | 0.003 | 0.340 | 62 | 0.9 | 0.01 |
| 0.297 | 0.323 | 0.996 | 0.004 | 0.357 | 63 | 0.9 | 0.01 |
| 0.323 | 0.350 | 0.996 | 0.004 | 0.374 | 64 | 0.9 | 0.01 |
| 0.350 | 0.377 | 0.996 | 0.004 | 0.393 | 65 | 0.9 | 0.01 |
| 0.377 | 0.404 | 0.996 | 0.004 | 0.413 | 66 | 0.9 | 0.01 |
| 0.404 | 0.431 | 0.996 | 0.004 | 0.434 | 67 | 0.9 | 0.01 |
| 0.431 | 0.459 | 0.995 | 0.005 | 0.456 | 68 | 0.9 | 0.01 |
| 0.459 | 0.487 | 0.995 | 0.005 | 0.480 | 69 | 0.9 | 0.01 |
| 0.487 | 0.515 | 0.995 | 0.005 | 0.506 | 70 | 0.9 | 0.01 |
| 0.515 | 0.544 | 0.995 | 0.005 | 0.533 | 71 | 0.9 | 0.01 |
| 0.544 | 0.574 | 0.994 | 0.006 | 0.562 | 72 | 0.9 | 0.01 |
| 0.574 | 0.604 | 0.994 | 0.006 | 0.594 | 73 | 0.9 | 0.01 |
| 0.604 | 0.634 | 0.994 | 0.006 | 0.628 | 74 | 0.9 | 0.01 |
| 0.634 | 0.665 | 0.993 | 0.007 | 0.664 | 75 | 0.9 | 0.01 |
| 0.665 | 0.697 | 0.993 | 0.007 | 0.703 | 76 | 0.9 | 0.01 |
| 0.697 | 0.730 | 0.993 | 0.007 | 0.746 | 77 | 0.9 | 0.01 |
| 0.730 | 0.763 | 0.992 | 0.008 | 0.793 | 78 | 0.9 | 0.01 |
| 0.763 | 0.798 | 0.992 | 0.008 | 0.843 | 79 | 0.9 | 0.01 |
| 0.798 | 0.833 | 0.991 | 0.009 | 0.898 | 80 | 0.9 | 0.01 |
| 0.833 | 0.869 | 0.990 | 0.010 | 0.959 | 81 | 0.9 | 0.01 |
| 0.869 | 0.907 | 0.990 | 0.010 | 1.026 | 82 | 0.9 | 0.01 |
| 0.907 | 0.946 | 0.989 | 0.011 | 1.101 | 83 | 0.9 | 0.01 |
| 0.946 | 0.987 | 0.988 | 0.012 | 1.183 | 84 | 0.9 | 0.01 |
| 0.987 | 1.029 | 0.987 | 0.013 | 1.276 | 85 | 0.9 | 0.01 |
| 1.029 | 1.073 | 0.986 | 0.014 | 1.381 | 86 | 0.9 | 0.01 |
| 1.073 | 1.120 | 0.985 | 0.015 | 1.499 | 87 | 0.9 | 0.01 |
| 1.120 | 1.169 | 0.984 | 0.016 | 1.635 | 88 | 0.9 | 0.01 |
| 1.169 | 1.222 | 0.982 | 0.018 | 1.793 | 89 | 0.9 | 0.01 |
| 1.222 | 1.278 | 0.980 | 0.020 | 1.977 | 90 | 0.9 | 0.01 |
| 1.278 | 1.338 | 0.978 | 0.022 | 2.196 | 91 | 0.9 | 0.01 |
| 1.338 | 1.404 | 0.975 | 0.025 | 2.459 | 92 | 0.9 | 0.01 |
| 1.404 | 1.476 | 0.972 | 0.028 | 2.783 | 93 | 0.9 | 0.01 |
| 1.476 | 1.558 | 0.968 | 0.032 | 3.193 | 94 | 0.9 | 0.01 |
| 1.558 | 1.651 | 0.963 | 0.037 | 3.728 | 95 | 0.9 | 0.01 |
| 1.651 | 1.761 | 0.955 | 0.045 | 4.459 | 96 | 0.9 | 0.01 |
| 1.761 | 1.898 | 0.945 | 0.055 | 5.529 | 97 | 0.9 | 0.01 |
| 1.898 | 2.082 | 0.927 | 0.073 | 7.271 | 98 | 0.9 | 0.01 |
| 2.082 | 2.380 | 0.892 | 0.108 | 10.768 | 99 | 0.9 | 0.01 |
| 2.380 | Inf | 0.734 | 0.266 | 26.623 | 100 | 0.9 | 0.01 |
| -Inf | -2.324 | 0.939 | 0.061 | 0.406 | 1 | 0.6 | 0.15 |
| -2.324 | -2.052 | 0.929 | 0.071 | 0.476 | 2 | 0.6 | 0.15 |
| -2.052 | -1.879 | 0.923 | 0.077 | 0.512 | 3 | 0.6 | 0.15 |
| -1.879 | -1.749 | 0.919 | 0.081 | 0.538 | 4 | 0.6 | 0.15 |
| -1.749 | -1.644 | 0.916 | 0.084 | 0.559 | 5 | 0.6 | 0.15 |
| -1.644 | -1.554 | 0.913 | 0.087 | 0.577 | 6 | 0.6 | 0.15 |
| -1.554 | -1.475 | 0.911 | 0.089 | 0.593 | 7 | 0.6 | 0.15 |
| -1.475 | -1.404 | 0.909 | 0.091 | 0.608 | 8 | 0.6 | 0.15 |
| -1.404 | -1.340 | 0.907 | 0.093 | 0.621 | 9 | 0.6 | 0.15 |
| -1.340 | -1.281 | 0.905 | 0.095 | 0.634 | 10 | 0.6 | 0.15 |
| -1.281 | -1.226 | 0.903 | 0.097 | 0.646 | 11 | 0.6 | 0.15 |
| -1.226 | -1.175 | 0.901 | 0.099 | 0.657 | 12 | 0.6 | 0.15 |
| -1.175 | -1.126 | 0.900 | 0.100 | 0.668 | 13 | 0.6 | 0.15 |
| -1.126 | -1.080 | 0.898 | 0.102 | 0.678 | 14 | 0.6 | 0.15 |
| -1.080 | -1.036 | 0.897 | 0.103 | 0.688 | 15 | 0.6 | 0.15 |
| -1.036 | -0.994 | 0.895 | 0.105 | 0.698 | 16 | 0.6 | 0.15 |
| -0.994 | -0.954 | 0.894 | 0.106 | 0.707 | 17 | 0.6 | 0.15 |
| -0.954 | -0.915 | 0.893 | 0.107 | 0.716 | 18 | 0.6 | 0.15 |
| -0.915 | -0.878 | 0.891 | 0.109 | 0.725 | 19 | 0.6 | 0.15 |
| -0.878 | -0.842 | 0.890 | 0.110 | 0.734 | 20 | 0.6 | 0.15 |
| -0.842 | -0.807 | 0.889 | 0.111 | 0.742 | 21 | 0.6 | 0.15 |
| -0.807 | -0.772 | 0.887 | 0.113 | 0.751 | 22 | 0.6 | 0.15 |
| -0.772 | -0.739 | 0.886 | 0.114 | 0.759 | 23 | 0.6 | 0.15 |
| -0.739 | -0.707 | 0.885 | 0.115 | 0.767 | 24 | 0.6 | 0.15 |
| -0.707 | -0.675 | 0.884 | 0.116 | 0.775 | 25 | 0.6 | 0.15 |
| -0.675 | -0.644 | 0.883 | 0.117 | 0.782 | 26 | 0.6 | 0.15 |
| -0.644 | -0.613 | 0.881 | 0.119 | 0.790 | 27 | 0.6 | 0.15 |
| -0.613 | -0.583 | 0.880 | 0.120 | 0.798 | 28 | 0.6 | 0.15 |
| -0.583 | -0.554 | 0.879 | 0.121 | 0.805 | 29 | 0.6 | 0.15 |
| -0.554 | -0.525 | 0.878 | 0.122 | 0.813 | 30 | 0.6 | 0.15 |
| -0.525 | -0.496 | 0.877 | 0.123 | 0.820 | 31 | 0.6 | 0.15 |
| -0.496 | -0.468 | 0.876 | 0.124 | 0.828 | 32 | 0.6 | 0.15 |
| -0.468 | -0.440 | 0.875 | 0.125 | 0.835 | 33 | 0.6 | 0.15 |
| -0.440 | -0.413 | 0.874 | 0.126 | 0.842 | 34 | 0.6 | 0.15 |
| -0.413 | -0.386 | 0.873 | 0.127 | 0.850 | 35 | 0.6 | 0.15 |
| -0.386 | -0.359 | 0.871 | 0.129 | 0.857 | 36 | 0.6 | 0.15 |
| -0.359 | -0.332 | 0.870 | 0.130 | 0.864 | 37 | 0.6 | 0.15 |
| -0.332 | -0.306 | 0.869 | 0.131 | 0.871 | 38 | 0.6 | 0.15 |
| -0.306 | -0.280 | 0.868 | 0.132 | 0.879 | 39 | 0.6 | 0.15 |
| -0.280 | -0.254 | 0.867 | 0.133 | 0.886 | 40 | 0.6 | 0.15 |
| -0.254 | -0.228 | 0.866 | 0.134 | 0.893 | 41 | 0.6 | 0.15 |
| -0.228 | -0.203 | 0.865 | 0.135 | 0.900 | 42 | 0.6 | 0.15 |
| -0.203 | -0.177 | 0.864 | 0.136 | 0.907 | 43 | 0.6 | 0.15 |
| -0.177 | -0.152 | 0.863 | 0.137 | 0.915 | 44 | 0.6 | 0.15 |
| -0.152 | -0.126 | 0.862 | 0.138 | 0.922 | 45 | 0.6 | 0.15 |
| -0.126 | -0.101 | 0.861 | 0.139 | 0.929 | 46 | 0.6 | 0.15 |
| -0.101 | -0.076 | 0.860 | 0.140 | 0.936 | 47 | 0.6 | 0.15 |
| -0.076 | -0.051 | 0.858 | 0.142 | 0.944 | 48 | 0.6 | 0.15 |
| -0.051 | -0.026 | 0.857 | 0.143 | 0.951 | 49 | 0.6 | 0.15 |
| -0.026 | -0.001 | 0.856 | 0.144 | 0.958 | 50 | 0.6 | 0.15 |
| -0.001 | 0.024 | 0.855 | 0.145 | 0.966 | 51 | 0.6 | 0.15 |
| 0.024 | 0.049 | 0.854 | 0.146 | 0.973 | 52 | 0.6 | 0.15 |
| 0.049 | 0.075 | 0.853 | 0.147 | 0.981 | 53 | 0.6 | 0.15 |
| 0.075 | 0.100 | 0.852 | 0.148 | 0.989 | 54 | 0.6 | 0.15 |
| 0.100 | 0.125 | 0.851 | 0.149 | 0.996 | 55 | 0.6 | 0.15 |
| 0.125 | 0.150 | 0.849 | 0.151 | 1.004 | 56 | 0.6 | 0.15 |
| 0.150 | 0.176 | 0.848 | 0.152 | 1.012 | 57 | 0.6 | 0.15 |
| 0.176 | 0.201 | 0.847 | 0.153 | 1.020 | 58 | 0.6 | 0.15 |
| 0.201 | 0.227 | 0.846 | 0.154 | 1.028 | 59 | 0.6 | 0.15 |
| 0.227 | 0.253 | 0.845 | 0.155 | 1.036 | 60 | 0.6 | 0.15 |
| 0.253 | 0.279 | 0.843 | 0.157 | 1.044 | 61 | 0.6 | 0.15 |
| 0.279 | 0.305 | 0.842 | 0.158 | 1.052 | 62 | 0.6 | 0.15 |
| 0.305 | 0.331 | 0.841 | 0.159 | 1.061 | 63 | 0.6 | 0.15 |
| 0.331 | 0.358 | 0.840 | 0.160 | 1.069 | 64 | 0.6 | 0.15 |
| 0.358 | 0.385 | 0.838 | 0.162 | 1.078 | 65 | 0.6 | 0.15 |
| 0.385 | 0.412 | 0.837 | 0.163 | 1.087 | 66 | 0.6 | 0.15 |
| 0.412 | 0.439 | 0.836 | 0.164 | 1.096 | 67 | 0.6 | 0.15 |
| 0.439 | 0.467 | 0.834 | 0.166 | 1.105 | 68 | 0.6 | 0.15 |
| 0.467 | 0.495 | 0.833 | 0.167 | 1.115 | 69 | 0.6 | 0.15 |
| 0.495 | 0.524 | 0.831 | 0.169 | 1.124 | 70 | 0.6 | 0.15 |
| 0.524 | 0.553 | 0.830 | 0.170 | 1.134 | 71 | 0.6 | 0.15 |
| 0.553 | 0.582 | 0.828 | 0.172 | 1.144 | 72 | 0.6 | 0.15 |
| 0.582 | 0.612 | 0.827 | 0.173 | 1.154 | 73 | 0.6 | 0.15 |
| 0.612 | 0.643 | 0.825 | 0.175 | 1.165 | 74 | 0.6 | 0.15 |
| 0.643 | 0.674 | 0.824 | 0.176 | 1.175 | 75 | 0.6 | 0.15 |
| 0.674 | 0.706 | 0.822 | 0.178 | 1.186 | 76 | 0.6 | 0.15 |
| 0.706 | 0.739 | 0.820 | 0.180 | 1.198 | 77 | 0.6 | 0.15 |
| 0.739 | 0.772 | 0.819 | 0.181 | 1.209 | 78 | 0.6 | 0.15 |
| 0.772 | 0.806 | 0.817 | 0.183 | 1.222 | 79 | 0.6 | 0.15 |
| 0.806 | 0.841 | 0.815 | 0.185 | 1.234 | 80 | 0.6 | 0.15 |
| 0.841 | 0.878 | 0.813 | 0.187 | 1.247 | 81 | 0.6 | 0.15 |
| 0.878 | 0.915 | 0.811 | 0.189 | 1.261 | 82 | 0.6 | 0.15 |
| 0.915 | 0.954 | 0.809 | 0.191 | 1.275 | 83 | 0.6 | 0.15 |
| 0.954 | 0.994 | 0.807 | 0.193 | 1.290 | 84 | 0.6 | 0.15 |
| 0.994 | 1.036 | 0.804 | 0.196 | 1.305 | 85 | 0.6 | 0.15 |
| 1.036 | 1.080 | 0.802 | 0.198 | 1.322 | 86 | 0.6 | 0.15 |
| 1.080 | 1.127 | 0.799 | 0.201 | 1.339 | 87 | 0.6 | 0.15 |
| 1.127 | 1.175 | 0.796 | 0.204 | 1.357 | 88 | 0.6 | 0.15 |
| 1.175 | 1.227 | 0.793 | 0.207 | 1.377 | 89 | 0.6 | 0.15 |
| 1.227 | 1.282 | 0.790 | 0.210 | 1.398 | 90 | 0.6 | 0.15 |
| 1.282 | 1.341 | 0.787 | 0.213 | 1.421 | 91 | 0.6 | 0.15 |
| 1.341 | 1.406 | 0.783 | 0.217 | 1.446 | 92 | 0.6 | 0.15 |
| 1.406 | 1.477 | 0.779 | 0.221 | 1.474 | 93 | 0.6 | 0.15 |
| 1.477 | 1.556 | 0.774 | 0.226 | 1.505 | 94 | 0.6 | 0.15 |
| 1.556 | 1.646 | 0.769 | 0.231 | 1.541 | 95 | 0.6 | 0.15 |
| 1.646 | 1.752 | 0.762 | 0.238 | 1.583 | 96 | 0.6 | 0.15 |
| 1.752 | 1.883 | 0.755 | 0.245 | 1.635 | 97 | 0.6 | 0.15 |
| 1.883 | 2.056 | 0.745 | 0.255 | 1.703 | 98 | 0.6 | 0.15 |
| 2.056 | 2.329 | 0.730 | 0.270 | 1.803 | 99 | 0.6 | 0.15 |
| 2.329 | Inf | 0.693 | 0.307 | 2.048 | 100 | 0.6 | 0.15 |
| -Inf | -2.304 | 0.981 | 0.019 | 0.129 | 1 | 0.7 | 0.15 |
| -2.304 | -2.037 | 0.973 | 0.027 | 0.181 | 2 | 0.7 | 0.15 |
| -2.037 | -1.867 | 0.968 | 0.032 | 0.211 | 3 | 0.7 | 0.15 |
| -1.867 | -1.739 | 0.965 | 0.035 | 0.235 | 4 | 0.7 | 0.15 |
| -1.739 | -1.635 | 0.962 | 0.038 | 0.256 | 5 | 0.7 | 0.15 |
| -1.635 | -1.546 | 0.959 | 0.041 | 0.275 | 6 | 0.7 | 0.15 |
| -1.546 | -1.469 | 0.956 | 0.044 | 0.292 | 7 | 0.7 | 0.15 |
| -1.469 | -1.399 | 0.954 | 0.046 | 0.308 | 8 | 0.7 | 0.15 |
| -1.399 | -1.336 | 0.951 | 0.049 | 0.324 | 9 | 0.7 | 0.15 |
| -1.336 | -1.277 | 0.949 | 0.051 | 0.338 | 10 | 0.7 | 0.15 |
| -1.277 | -1.223 | 0.947 | 0.053 | 0.352 | 11 | 0.7 | 0.15 |
| -1.223 | -1.172 | 0.945 | 0.055 | 0.366 | 12 | 0.7 | 0.15 |
| -1.172 | -1.124 | 0.943 | 0.057 | 0.379 | 13 | 0.7 | 0.15 |
| -1.124 | -1.079 | 0.941 | 0.059 | 0.392 | 14 | 0.7 | 0.15 |
| -1.079 | -1.035 | 0.939 | 0.061 | 0.405 | 15 | 0.7 | 0.15 |
| -1.035 | -0.994 | 0.937 | 0.063 | 0.418 | 16 | 0.7 | 0.15 |
| -0.994 | -0.954 | 0.935 | 0.065 | 0.430 | 17 | 0.7 | 0.15 |
| -0.954 | -0.915 | 0.934 | 0.066 | 0.442 | 18 | 0.7 | 0.15 |
| -0.915 | -0.878 | 0.932 | 0.068 | 0.454 | 19 | 0.7 | 0.15 |
| -0.878 | -0.842 | 0.930 | 0.070 | 0.466 | 20 | 0.7 | 0.15 |
| -0.842 | -0.808 | 0.928 | 0.072 | 0.478 | 21 | 0.7 | 0.15 |
| -0.808 | -0.774 | 0.926 | 0.074 | 0.490 | 22 | 0.7 | 0.15 |
| -0.774 | -0.741 | 0.925 | 0.075 | 0.502 | 23 | 0.7 | 0.15 |
| -0.741 | -0.708 | 0.923 | 0.077 | 0.514 | 24 | 0.7 | 0.15 |
| -0.708 | -0.677 | 0.921 | 0.079 | 0.526 | 25 | 0.7 | 0.15 |
| -0.677 | -0.646 | 0.919 | 0.081 | 0.537 | 26 | 0.7 | 0.15 |
| -0.646 | -0.616 | 0.918 | 0.082 | 0.549 | 27 | 0.7 | 0.15 |
| -0.616 | -0.586 | 0.916 | 0.084 | 0.561 | 28 | 0.7 | 0.15 |
| -0.586 | -0.557 | 0.914 | 0.086 | 0.572 | 29 | 0.7 | 0.15 |
| -0.557 | -0.528 | 0.912 | 0.088 | 0.584 | 30 | 0.7 | 0.15 |
| -0.528 | -0.500 | 0.911 | 0.089 | 0.596 | 31 | 0.7 | 0.15 |
| -0.500 | -0.472 | 0.909 | 0.091 | 0.608 | 32 | 0.7 | 0.15 |
| -0.472 | -0.444 | 0.907 | 0.093 | 0.620 | 33 | 0.7 | 0.15 |
| -0.444 | -0.417 | 0.905 | 0.095 | 0.632 | 34 | 0.7 | 0.15 |
| -0.417 | -0.390 | 0.903 | 0.097 | 0.644 | 35 | 0.7 | 0.15 |
| -0.390 | -0.363 | 0.902 | 0.098 | 0.656 | 36 | 0.7 | 0.15 |
| -0.363 | -0.337 | 0.900 | 0.100 | 0.668 | 37 | 0.7 | 0.15 |
| -0.337 | -0.310 | 0.898 | 0.102 | 0.680 | 38 | 0.7 | 0.15 |
| -0.310 | -0.284 | 0.896 | 0.104 | 0.692 | 39 | 0.7 | 0.15 |
| -0.284 | -0.258 | 0.894 | 0.106 | 0.705 | 40 | 0.7 | 0.15 |
| -0.258 | -0.233 | 0.892 | 0.108 | 0.717 | 41 | 0.7 | 0.15 |
| -0.233 | -0.207 | 0.890 | 0.110 | 0.730 | 42 | 0.7 | 0.15 |
| -0.207 | -0.182 | 0.889 | 0.111 | 0.743 | 43 | 0.7 | 0.15 |
| -0.182 | -0.156 | 0.887 | 0.113 | 0.756 | 44 | 0.7 | 0.15 |
| -0.156 | -0.131 | 0.885 | 0.115 | 0.769 | 45 | 0.7 | 0.15 |
| -0.131 | -0.106 | 0.883 | 0.117 | 0.782 | 46 | 0.7 | 0.15 |
| -0.106 | -0.081 | 0.881 | 0.119 | 0.796 | 47 | 0.7 | 0.15 |
| -0.081 | -0.056 | 0.879 | 0.121 | 0.809 | 48 | 0.7 | 0.15 |
| -0.056 | -0.031 | 0.877 | 0.123 | 0.823 | 49 | 0.7 | 0.15 |
| -0.031 | -0.006 | 0.874 | 0.126 | 0.837 | 50 | 0.7 | 0.15 |
| -0.006 | 0.019 | 0.872 | 0.128 | 0.851 | 51 | 0.7 | 0.15 |
| 0.019 | 0.044 | 0.870 | 0.130 | 0.865 | 52 | 0.7 | 0.15 |
| 0.044 | 0.070 | 0.868 | 0.132 | 0.880 | 53 | 0.7 | 0.15 |
| 0.070 | 0.095 | 0.866 | 0.134 | 0.895 | 54 | 0.7 | 0.15 |
| 0.095 | 0.120 | 0.864 | 0.136 | 0.910 | 55 | 0.7 | 0.15 |
| 0.120 | 0.145 | 0.861 | 0.139 | 0.925 | 56 | 0.7 | 0.15 |
| 0.145 | 0.171 | 0.859 | 0.141 | 0.941 | 57 | 0.7 | 0.15 |
| 0.171 | 0.196 | 0.856 | 0.144 | 0.957 | 58 | 0.7 | 0.15 |
| 0.196 | 0.222 | 0.854 | 0.146 | 0.973 | 59 | 0.7 | 0.15 |
| 0.222 | 0.248 | 0.852 | 0.148 | 0.990 | 60 | 0.7 | 0.15 |
| 0.248 | 0.274 | 0.849 | 0.151 | 1.006 | 61 | 0.7 | 0.15 |
| 0.274 | 0.300 | 0.846 | 0.154 | 1.024 | 62 | 0.7 | 0.15 |
| 0.300 | 0.326 | 0.844 | 0.156 | 1.041 | 63 | 0.7 | 0.15 |
| 0.326 | 0.353 | 0.841 | 0.159 | 1.059 | 64 | 0.7 | 0.15 |
| 0.353 | 0.380 | 0.838 | 0.162 | 1.078 | 65 | 0.7 | 0.15 |
| 0.380 | 0.407 | 0.835 | 0.165 | 1.097 | 66 | 0.7 | 0.15 |
| 0.407 | 0.435 | 0.833 | 0.167 | 1.116 | 67 | 0.7 | 0.15 |
| 0.435 | 0.463 | 0.830 | 0.170 | 1.136 | 68 | 0.7 | 0.15 |
| 0.463 | 0.491 | 0.827 | 0.173 | 1.156 | 69 | 0.7 | 0.15 |
| 0.491 | 0.520 | 0.823 | 0.177 | 1.178 | 70 | 0.7 | 0.15 |
| 0.520 | 0.549 | 0.820 | 0.180 | 1.199 | 71 | 0.7 | 0.15 |
| 0.549 | 0.579 | 0.817 | 0.183 | 1.222 | 72 | 0.7 | 0.15 |
| 0.579 | 0.609 | 0.813 | 0.187 | 1.245 | 73 | 0.7 | 0.15 |
| 0.609 | 0.639 | 0.810 | 0.190 | 1.268 | 74 | 0.7 | 0.15 |
| 0.639 | 0.671 | 0.806 | 0.194 | 1.293 | 75 | 0.7 | 0.15 |
| 0.671 | 0.703 | 0.802 | 0.198 | 1.319 | 76 | 0.7 | 0.15 |
| 0.703 | 0.736 | 0.798 | 0.202 | 1.345 | 77 | 0.7 | 0.15 |
| 0.736 | 0.769 | 0.794 | 0.206 | 1.373 | 78 | 0.7 | 0.15 |
| 0.769 | 0.804 | 0.790 | 0.210 | 1.401 | 79 | 0.7 | 0.15 |
| 0.804 | 0.839 | 0.785 | 0.215 | 1.431 | 80 | 0.7 | 0.15 |
| 0.839 | 0.876 | 0.781 | 0.219 | 1.463 | 81 | 0.7 | 0.15 |
| 0.876 | 0.914 | 0.776 | 0.224 | 1.496 | 82 | 0.7 | 0.15 |
| 0.914 | 0.953 | 0.770 | 0.230 | 1.530 | 83 | 0.7 | 0.15 |
| 0.953 | 0.994 | 0.765 | 0.235 | 1.567 | 84 | 0.7 | 0.15 |
| 0.994 | 1.036 | 0.759 | 0.241 | 1.605 | 85 | 0.7 | 0.15 |
| 1.036 | 1.081 | 0.753 | 0.247 | 1.646 | 86 | 0.7 | 0.15 |
| 1.081 | 1.127 | 0.747 | 0.253 | 1.690 | 87 | 0.7 | 0.15 |
| 1.127 | 1.177 | 0.740 | 0.260 | 1.737 | 88 | 0.7 | 0.15 |
| 1.177 | 1.229 | 0.732 | 0.268 | 1.787 | 89 | 0.7 | 0.15 |
| 1.229 | 1.285 | 0.724 | 0.276 | 1.842 | 90 | 0.7 | 0.15 |
| 1.285 | 1.345 | 0.715 | 0.285 | 1.902 | 91 | 0.7 | 0.15 |
| 1.345 | 1.410 | 0.705 | 0.295 | 1.968 | 92 | 0.7 | 0.15 |
| 1.410 | 1.482 | 0.694 | 0.306 | 2.041 | 93 | 0.7 | 0.15 |
| 1.482 | 1.563 | 0.681 | 0.319 | 2.125 | 94 | 0.7 | 0.15 |
| 1.563 | 1.655 | 0.667 | 0.333 | 2.221 | 95 | 0.7 | 0.15 |
| 1.655 | 1.763 | 0.650 | 0.350 | 2.336 | 96 | 0.7 | 0.15 |
| 1.763 | 1.896 | 0.628 | 0.372 | 2.477 | 97 | 0.7 | 0.15 |
| 1.896 | 2.073 | 0.601 | 0.399 | 2.662 | 98 | 0.7 | 0.15 |
| 2.073 | 2.352 | 0.560 | 0.440 | 2.935 | 99 | 0.7 | 0.15 |
| 2.352 | Inf | 0.465 | 0.535 | 3.568 | 100 | 0.7 | 0.15 |
| -Inf | -2.250 | 0.996 | 0.004 | 0.027 | 1 | 0.8 | 0.15 |
| -2.250 | -1.994 | 0.993 | 0.007 | 0.047 | 2 | 0.8 | 0.15 |
| -1.994 | -1.831 | 0.991 | 0.009 | 0.060 | 3 | 0.8 | 0.15 |
| -1.831 | -1.708 | 0.989 | 0.011 | 0.072 | 4 | 0.8 | 0.15 |
| -1.708 | -1.608 | 0.988 | 0.012 | 0.083 | 5 | 0.8 | 0.15 |
| -1.608 | -1.523 | 0.986 | 0.014 | 0.093 | 6 | 0.8 | 0.15 |
| -1.523 | -1.448 | 0.984 | 0.016 | 0.103 | 7 | 0.8 | 0.15 |
| -1.448 | -1.381 | 0.983 | 0.017 | 0.113 | 8 | 0.8 | 0.15 |
| -1.381 | -1.320 | 0.982 | 0.018 | 0.123 | 9 | 0.8 | 0.15 |
| -1.320 | -1.264 | 0.980 | 0.020 | 0.132 | 10 | 0.8 | 0.15 |
| -1.264 | -1.211 | 0.979 | 0.021 | 0.142 | 11 | 0.8 | 0.15 |
| -1.211 | -1.162 | 0.977 | 0.023 | 0.151 | 12 | 0.8 | 0.15 |
| -1.162 | -1.116 | 0.976 | 0.024 | 0.160 | 13 | 0.8 | 0.15 |
| -1.116 | -1.072 | 0.975 | 0.025 | 0.170 | 14 | 0.8 | 0.15 |
| -1.072 | -1.030 | 0.973 | 0.027 | 0.179 | 15 | 0.8 | 0.15 |
| -1.030 | -0.989 | 0.972 | 0.028 | 0.189 | 16 | 0.8 | 0.15 |
| -0.989 | -0.951 | 0.970 | 0.030 | 0.198 | 17 | 0.8 | 0.15 |
| -0.951 | -0.914 | 0.969 | 0.031 | 0.208 | 18 | 0.8 | 0.15 |
| -0.914 | -0.878 | 0.967 | 0.033 | 0.218 | 19 | 0.8 | 0.15 |
| -0.878 | -0.843 | 0.966 | 0.034 | 0.228 | 20 | 0.8 | 0.15 |
| -0.843 | -0.809 | 0.964 | 0.036 | 0.238 | 21 | 0.8 | 0.15 |
| -0.809 | -0.776 | 0.963 | 0.037 | 0.248 | 22 | 0.8 | 0.15 |
| -0.776 | -0.744 | 0.961 | 0.039 | 0.258 | 23 | 0.8 | 0.15 |
| -0.744 | -0.712 | 0.960 | 0.040 | 0.268 | 24 | 0.8 | 0.15 |
| -0.712 | -0.681 | 0.958 | 0.042 | 0.279 | 25 | 0.8 | 0.15 |
| -0.681 | -0.651 | 0.957 | 0.043 | 0.290 | 26 | 0.8 | 0.15 |
| -0.651 | -0.622 | 0.955 | 0.045 | 0.300 | 27 | 0.8 | 0.15 |
| -0.622 | -0.593 | 0.953 | 0.047 | 0.311 | 28 | 0.8 | 0.15 |
| -0.593 | -0.564 | 0.952 | 0.048 | 0.323 | 29 | 0.8 | 0.15 |
| -0.564 | -0.536 | 0.950 | 0.050 | 0.334 | 30 | 0.8 | 0.15 |
| -0.536 | -0.508 | 0.948 | 0.052 | 0.346 | 31 | 0.8 | 0.15 |
| -0.508 | -0.481 | 0.946 | 0.054 | 0.358 | 32 | 0.8 | 0.15 |
| -0.481 | -0.454 | 0.945 | 0.055 | 0.370 | 33 | 0.8 | 0.15 |
| -0.454 | -0.427 | 0.943 | 0.057 | 0.382 | 34 | 0.8 | 0.15 |
| -0.427 | -0.401 | 0.941 | 0.059 | 0.395 | 35 | 0.8 | 0.15 |
| -0.401 | -0.374 | 0.939 | 0.061 | 0.408 | 36 | 0.8 | 0.15 |
| -0.374 | -0.348 | 0.937 | 0.063 | 0.421 | 37 | 0.8 | 0.15 |
| -0.348 | -0.322 | 0.935 | 0.065 | 0.434 | 38 | 0.8 | 0.15 |
| -0.322 | -0.297 | 0.933 | 0.067 | 0.448 | 39 | 0.8 | 0.15 |
| -0.297 | -0.271 | 0.931 | 0.069 | 0.462 | 40 | 0.8 | 0.15 |
| -0.271 | -0.246 | 0.929 | 0.071 | 0.476 | 41 | 0.8 | 0.15 |
| -0.246 | -0.221 | 0.926 | 0.074 | 0.491 | 42 | 0.8 | 0.15 |
| -0.221 | -0.196 | 0.924 | 0.076 | 0.506 | 43 | 0.8 | 0.15 |
| -0.196 | -0.171 | 0.922 | 0.078 | 0.521 | 44 | 0.8 | 0.15 |
| -0.171 | -0.146 | 0.919 | 0.081 | 0.537 | 45 | 0.8 | 0.15 |
| -0.146 | -0.121 | 0.917 | 0.083 | 0.553 | 46 | 0.8 | 0.15 |
| -0.121 | -0.096 | 0.915 | 0.085 | 0.570 | 47 | 0.8 | 0.15 |
| -0.096 | -0.071 | 0.912 | 0.088 | 0.587 | 48 | 0.8 | 0.15 |
| -0.071 | -0.046 | 0.909 | 0.091 | 0.604 | 49 | 0.8 | 0.15 |
| -0.046 | -0.021 | 0.907 | 0.093 | 0.622 | 50 | 0.8 | 0.15 |
| -0.021 | 0.003 | 0.904 | 0.096 | 0.640 | 51 | 0.8 | 0.15 |
| 0.003 | 0.028 | 0.901 | 0.099 | 0.659 | 52 | 0.8 | 0.15 |
| 0.028 | 0.053 | 0.898 | 0.102 | 0.679 | 53 | 0.8 | 0.15 |
| 0.053 | 0.078 | 0.895 | 0.105 | 0.699 | 54 | 0.8 | 0.15 |
| 0.078 | 0.104 | 0.892 | 0.108 | 0.719 | 55 | 0.8 | 0.15 |
| 0.104 | 0.129 | 0.889 | 0.111 | 0.740 | 56 | 0.8 | 0.15 |
| 0.129 | 0.154 | 0.886 | 0.114 | 0.762 | 57 | 0.8 | 0.15 |
| 0.154 | 0.180 | 0.882 | 0.118 | 0.785 | 58 | 0.8 | 0.15 |
| 0.180 | 0.206 | 0.879 | 0.121 | 0.808 | 59 | 0.8 | 0.15 |
| 0.206 | 0.231 | 0.875 | 0.125 | 0.832 | 60 | 0.8 | 0.15 |
| 0.231 | 0.258 | 0.871 | 0.129 | 0.857 | 61 | 0.8 | 0.15 |
| 0.258 | 0.284 | 0.868 | 0.132 | 0.882 | 62 | 0.8 | 0.15 |
| 0.284 | 0.311 | 0.864 | 0.136 | 0.909 | 63 | 0.8 | 0.15 |
| 0.311 | 0.337 | 0.860 | 0.140 | 0.936 | 64 | 0.8 | 0.15 |
| 0.337 | 0.365 | 0.855 | 0.145 | 0.965 | 65 | 0.8 | 0.15 |
| 0.365 | 0.392 | 0.851 | 0.149 | 0.994 | 66 | 0.8 | 0.15 |
| 0.392 | 0.420 | 0.846 | 0.154 | 1.025 | 67 | 0.8 | 0.15 |
| 0.420 | 0.448 | 0.841 | 0.159 | 1.057 | 68 | 0.8 | 0.15 |
| 0.448 | 0.477 | 0.837 | 0.163 | 1.090 | 69 | 0.8 | 0.15 |
| 0.477 | 0.506 | 0.831 | 0.169 | 1.124 | 70 | 0.8 | 0.15 |
| 0.506 | 0.535 | 0.826 | 0.174 | 1.160 | 71 | 0.8 | 0.15 |
| 0.535 | 0.565 | 0.820 | 0.180 | 1.198 | 72 | 0.8 | 0.15 |
| 0.565 | 0.596 | 0.814 | 0.186 | 1.237 | 73 | 0.8 | 0.15 |
| 0.596 | 0.627 | 0.808 | 0.192 | 1.278 | 74 | 0.8 | 0.15 |
| 0.627 | 0.659 | 0.802 | 0.198 | 1.320 | 75 | 0.8 | 0.15 |
| 0.659 | 0.692 | 0.795 | 0.205 | 1.365 | 76 | 0.8 | 0.15 |
| 0.692 | 0.726 | 0.788 | 0.212 | 1.412 | 77 | 0.8 | 0.15 |
| 0.726 | 0.760 | 0.781 | 0.219 | 1.462 | 78 | 0.8 | 0.15 |
| 0.760 | 0.796 | 0.773 | 0.227 | 1.514 | 79 | 0.8 | 0.15 |
| 0.796 | 0.832 | 0.765 | 0.235 | 1.569 | 80 | 0.8 | 0.15 |
| 0.832 | 0.870 | 0.756 | 0.244 | 1.628 | 81 | 0.8 | 0.15 |
| 0.870 | 0.909 | 0.747 | 0.253 | 1.689 | 82 | 0.8 | 0.15 |
| 0.909 | 0.949 | 0.737 | 0.263 | 1.755 | 83 | 0.8 | 0.15 |
| 0.949 | 0.991 | 0.726 | 0.274 | 1.825 | 84 | 0.8 | 0.15 |
| 0.991 | 1.035 | 0.715 | 0.285 | 1.900 | 85 | 0.8 | 0.15 |
| 1.035 | 1.081 | 0.703 | 0.297 | 1.980 | 86 | 0.8 | 0.15 |
| 1.081 | 1.130 | 0.690 | 0.310 | 2.066 | 87 | 0.8 | 0.15 |
| 1.130 | 1.181 | 0.676 | 0.324 | 2.159 | 88 | 0.8 | 0.15 |
| 1.181 | 1.236 | 0.661 | 0.339 | 2.260 | 89 | 0.8 | 0.15 |
| 1.236 | 1.294 | 0.644 | 0.356 | 2.370 | 90 | 0.8 | 0.15 |
| 1.294 | 1.357 | 0.626 | 0.374 | 2.492 | 91 | 0.8 | 0.15 |
| 1.357 | 1.426 | 0.606 | 0.394 | 2.626 | 92 | 0.8 | 0.15 |
| 1.426 | 1.502 | 0.584 | 0.416 | 2.775 | 93 | 0.8 | 0.15 |
| 1.502 | 1.587 | 0.558 | 0.442 | 2.944 | 94 | 0.8 | 0.15 |
| 1.587 | 1.684 | 0.529 | 0.471 | 3.138 | 95 | 0.8 | 0.15 |
| 1.684 | 1.798 | 0.495 | 0.505 | 3.364 | 96 | 0.8 | 0.15 |
| 1.798 | 1.939 | 0.455 | 0.545 | 3.636 | 97 | 0.8 | 0.15 |
| 1.939 | 2.126 | 0.404 | 0.596 | 3.976 | 98 | 0.8 | 0.15 |
| 2.126 | 2.422 | 0.334 | 0.666 | 4.439 | 99 | 0.8 | 0.15 |
| 2.422 | Inf | 0.211 | 0.789 | 5.263 | 100 | 0.8 | 0.15 |
| -Inf | -2.130 | 1.000 | 0.000 | 0.002 | 1 | 0.9 | 0.15 |
| -2.130 | -1.896 | 0.999 | 0.001 | 0.005 | 2 | 0.9 | 0.15 |
| -1.896 | -1.746 | 0.999 | 0.001 | 0.007 | 3 | 0.9 | 0.15 |
| -1.746 | -1.634 | 0.999 | 0.001 | 0.010 | 4 | 0.9 | 0.15 |
| -1.634 | -1.542 | 0.998 | 0.002 | 0.012 | 5 | 0.9 | 0.15 |
| -1.542 | -1.464 | 0.998 | 0.002 | 0.015 | 6 | 0.9 | 0.15 |
| -1.464 | -1.395 | 0.997 | 0.003 | 0.017 | 7 | 0.9 | 0.15 |
| -1.395 | -1.334 | 0.997 | 0.003 | 0.020 | 8 | 0.9 | 0.15 |
| -1.334 | -1.277 | 0.997 | 0.003 | 0.022 | 9 | 0.9 | 0.15 |
| -1.277 | -1.226 | 0.996 | 0.004 | 0.025 | 10 | 0.9 | 0.15 |
| -1.226 | -1.177 | 0.996 | 0.004 | 0.028 | 11 | 0.9 | 0.15 |
| -1.177 | -1.132 | 0.995 | 0.005 | 0.031 | 12 | 0.9 | 0.15 |
| -1.132 | -1.089 | 0.995 | 0.005 | 0.034 | 13 | 0.9 | 0.15 |
| -1.089 | -1.049 | 0.994 | 0.006 | 0.037 | 14 | 0.9 | 0.15 |
| -1.049 | -1.010 | 0.994 | 0.006 | 0.040 | 15 | 0.9 | 0.15 |
| -1.010 | -0.973 | 0.993 | 0.007 | 0.044 | 16 | 0.9 | 0.15 |
| -0.973 | -0.937 | 0.993 | 0.007 | 0.047 | 17 | 0.9 | 0.15 |
| -0.937 | -0.902 | 0.992 | 0.008 | 0.051 | 18 | 0.9 | 0.15 |
| -0.902 | -0.869 | 0.992 | 0.008 | 0.055 | 19 | 0.9 | 0.15 |
| -0.869 | -0.837 | 0.991 | 0.009 | 0.059 | 20 | 0.9 | 0.15 |
| -0.837 | -0.805 | 0.991 | 0.009 | 0.063 | 21 | 0.9 | 0.15 |
| -0.805 | -0.775 | 0.990 | 0.010 | 0.067 | 22 | 0.9 | 0.15 |
| -0.775 | -0.745 | 0.989 | 0.011 | 0.072 | 23 | 0.9 | 0.15 |
| -0.745 | -0.715 | 0.989 | 0.011 | 0.076 | 24 | 0.9 | 0.15 |
| -0.715 | -0.687 | 0.988 | 0.012 | 0.081 | 25 | 0.9 | 0.15 |
| -0.687 | -0.659 | 0.987 | 0.013 | 0.086 | 26 | 0.9 | 0.15 |
| -0.659 | -0.631 | 0.986 | 0.014 | 0.091 | 27 | 0.9 | 0.15 |
| -0.631 | -0.604 | 0.985 | 0.015 | 0.097 | 28 | 0.9 | 0.15 |
| -0.604 | -0.577 | 0.985 | 0.015 | 0.103 | 29 | 0.9 | 0.15 |
| -0.577 | -0.551 | 0.984 | 0.016 | 0.109 | 30 | 0.9 | 0.15 |
| -0.551 | -0.525 | 0.983 | 0.017 | 0.115 | 31 | 0.9 | 0.15 |
| -0.525 | -0.499 | 0.982 | 0.018 | 0.121 | 32 | 0.9 | 0.15 |
| -0.499 | -0.473 | 0.981 | 0.019 | 0.128 | 33 | 0.9 | 0.15 |
| -0.473 | -0.448 | 0.980 | 0.020 | 0.135 | 34 | 0.9 | 0.15 |
| -0.448 | -0.423 | 0.979 | 0.021 | 0.142 | 35 | 0.9 | 0.15 |
| -0.423 | -0.398 | 0.977 | 0.023 | 0.150 | 36 | 0.9 | 0.15 |
| -0.398 | -0.374 | 0.976 | 0.024 | 0.158 | 37 | 0.9 | 0.15 |
| -0.374 | -0.349 | 0.975 | 0.025 | 0.166 | 38 | 0.9 | 0.15 |
| -0.349 | -0.325 | 0.974 | 0.026 | 0.175 | 39 | 0.9 | 0.15 |
| -0.325 | -0.301 | 0.972 | 0.028 | 0.184 | 40 | 0.9 | 0.15 |
| -0.301 | -0.277 | 0.971 | 0.029 | 0.194 | 41 | 0.9 | 0.15 |
| -0.277 | -0.253 | 0.969 | 0.031 | 0.204 | 42 | 0.9 | 0.15 |
| -0.253 | -0.229 | 0.968 | 0.032 | 0.214 | 43 | 0.9 | 0.15 |
| -0.229 | -0.205 | 0.966 | 0.034 | 0.225 | 44 | 0.9 | 0.15 |
| -0.205 | -0.181 | 0.964 | 0.036 | 0.237 | 45 | 0.9 | 0.15 |
| -0.181 | -0.157 | 0.963 | 0.037 | 0.249 | 46 | 0.9 | 0.15 |
| -0.157 | -0.133 | 0.961 | 0.039 | 0.262 | 47 | 0.9 | 0.15 |
| -0.133 | -0.109 | 0.959 | 0.041 | 0.275 | 48 | 0.9 | 0.15 |
| -0.109 | -0.085 | 0.957 | 0.043 | 0.289 | 49 | 0.9 | 0.15 |
| -0.085 | -0.061 | 0.954 | 0.046 | 0.304 | 50 | 0.9 | 0.15 |
| -0.061 | -0.037 | 0.952 | 0.048 | 0.319 | 51 | 0.9 | 0.15 |
| -0.037 | -0.013 | 0.950 | 0.050 | 0.335 | 52 | 0.9 | 0.15 |
| -0.013 | 0.011 | 0.947 | 0.053 | 0.352 | 53 | 0.9 | 0.15 |
| 0.011 | 0.036 | 0.944 | 0.056 | 0.370 | 54 | 0.9 | 0.15 |
| 0.036 | 0.060 | 0.942 | 0.058 | 0.389 | 55 | 0.9 | 0.15 |
| 0.060 | 0.085 | 0.939 | 0.061 | 0.409 | 56 | 0.9 | 0.15 |
| 0.085 | 0.110 | 0.935 | 0.065 | 0.430 | 57 | 0.9 | 0.15 |
| 0.110 | 0.135 | 0.932 | 0.068 | 0.453 | 58 | 0.9 | 0.15 |
| 0.135 | 0.161 | 0.929 | 0.071 | 0.476 | 59 | 0.9 | 0.15 |
| 0.161 | 0.187 | 0.925 | 0.075 | 0.501 | 60 | 0.9 | 0.15 |
| 0.187 | 0.212 | 0.921 | 0.079 | 0.528 | 61 | 0.9 | 0.15 |
| 0.212 | 0.239 | 0.917 | 0.083 | 0.556 | 62 | 0.9 | 0.15 |
| 0.239 | 0.265 | 0.912 | 0.088 | 0.586 | 63 | 0.9 | 0.15 |
| 0.265 | 0.292 | 0.907 | 0.093 | 0.617 | 64 | 0.9 | 0.15 |
| 0.292 | 0.320 | 0.902 | 0.098 | 0.651 | 65 | 0.9 | 0.15 |
| 0.320 | 0.347 | 0.897 | 0.103 | 0.687 | 66 | 0.9 | 0.15 |
| 0.347 | 0.376 | 0.891 | 0.109 | 0.725 | 67 | 0.9 | 0.15 |
| 0.376 | 0.404 | 0.885 | 0.115 | 0.766 | 68 | 0.9 | 0.15 |
| 0.404 | 0.434 | 0.879 | 0.121 | 0.809 | 69 | 0.9 | 0.15 |
| 0.434 | 0.464 | 0.872 | 0.128 | 0.856 | 70 | 0.9 | 0.15 |
| 0.464 | 0.494 | 0.864 | 0.136 | 0.905 | 71 | 0.9 | 0.15 |
| 0.494 | 0.525 | 0.856 | 0.144 | 0.959 | 72 | 0.9 | 0.15 |
| 0.525 | 0.557 | 0.848 | 0.152 | 1.016 | 73 | 0.9 | 0.15 |
| 0.557 | 0.590 | 0.838 | 0.162 | 1.078 | 74 | 0.9 | 0.15 |
| 0.590 | 0.624 | 0.828 | 0.172 | 1.144 | 75 | 0.9 | 0.15 |
| 0.624 | 0.658 | 0.818 | 0.182 | 1.215 | 76 | 0.9 | 0.15 |
| 0.658 | 0.694 | 0.806 | 0.194 | 1.293 | 77 | 0.9 | 0.15 |
| 0.694 | 0.731 | 0.794 | 0.206 | 1.376 | 78 | 0.9 | 0.15 |
| 0.731 | 0.769 | 0.780 | 0.220 | 1.466 | 79 | 0.9 | 0.15 |
| 0.769 | 0.808 | 0.765 | 0.235 | 1.564 | 80 | 0.9 | 0.15 |
| 0.808 | 0.849 | 0.749 | 0.251 | 1.670 | 81 | 0.9 | 0.15 |
| 0.849 | 0.892 | 0.732 | 0.268 | 1.786 | 82 | 0.9 | 0.15 |
| 0.892 | 0.936 | 0.713 | 0.287 | 1.912 | 83 | 0.9 | 0.15 |
| 0.936 | 0.983 | 0.693 | 0.307 | 2.049 | 84 | 0.9 | 0.15 |
| 0.983 | 1.032 | 0.670 | 0.330 | 2.198 | 85 | 0.9 | 0.15 |
| 1.032 | 1.084 | 0.646 | 0.354 | 2.361 | 86 | 0.9 | 0.15 |
| 1.084 | 1.138 | 0.619 | 0.381 | 2.538 | 87 | 0.9 | 0.15 |
| 1.138 | 1.197 | 0.590 | 0.410 | 2.732 | 88 | 0.9 | 0.15 |
| 1.197 | 1.259 | 0.558 | 0.442 | 2.944 | 89 | 0.9 | 0.15 |
| 1.259 | 1.325 | 0.524 | 0.476 | 3.174 | 90 | 0.9 | 0.15 |
| 1.325 | 1.398 | 0.486 | 0.514 | 3.424 | 91 | 0.9 | 0.15 |
| 1.398 | 1.477 | 0.446 | 0.554 | 3.694 | 92 | 0.9 | 0.15 |
| 1.477 | 1.564 | 0.402 | 0.598 | 3.984 | 93 | 0.9 | 0.15 |
| 1.564 | 1.661 | 0.356 | 0.644 | 4.295 | 94 | 0.9 | 0.15 |
| 1.661 | 1.771 | 0.306 | 0.694 | 4.624 | 95 | 0.9 | 0.15 |
| 1.771 | 1.900 | 0.255 | 0.745 | 4.968 | 96 | 0.9 | 0.15 |
| 1.900 | 2.057 | 0.201 | 0.799 | 5.324 | 97 | 0.9 | 0.15 |
| 2.057 | 2.260 | 0.147 | 0.853 | 5.687 | 98 | 0.9 | 0.15 |
| 2.260 | 2.570 | 0.092 | 0.908 | 6.050 | 99 | 0.9 | 0.15 |
| 2.570 | Inf | 0.037 | 0.963 | 6.423 | 100 | 0.9 | 0.15 |
| -Inf | -2.323 | 0.862 | 0.138 | 0.459 | 1 | 0.6 | 0.30 |
| -2.323 | -2.052 | 0.841 | 0.159 | 0.532 | 2 | 0.6 | 0.30 |
| -2.052 | -1.879 | 0.830 | 0.170 | 0.567 | 3 | 0.6 | 0.30 |
| -1.879 | -1.749 | 0.822 | 0.178 | 0.593 | 4 | 0.6 | 0.30 |
| -1.749 | -1.644 | 0.816 | 0.184 | 0.614 | 5 | 0.6 | 0.30 |
| -1.644 | -1.554 | 0.810 | 0.190 | 0.632 | 6 | 0.6 | 0.30 |
| -1.554 | -1.475 | 0.806 | 0.194 | 0.648 | 7 | 0.6 | 0.30 |
| -1.475 | -1.404 | 0.801 | 0.199 | 0.662 | 8 | 0.6 | 0.30 |
| -1.404 | -1.340 | 0.798 | 0.202 | 0.675 | 9 | 0.6 | 0.30 |
| -1.340 | -1.281 | 0.794 | 0.206 | 0.687 | 10 | 0.6 | 0.30 |
| -1.281 | -1.226 | 0.791 | 0.209 | 0.698 | 11 | 0.6 | 0.30 |
| -1.226 | -1.175 | 0.787 | 0.213 | 0.709 | 12 | 0.6 | 0.30 |
| -1.175 | -1.126 | 0.784 | 0.216 | 0.719 | 13 | 0.6 | 0.30 |
| -1.126 | -1.080 | 0.781 | 0.219 | 0.729 | 14 | 0.6 | 0.30 |
| -1.080 | -1.036 | 0.779 | 0.221 | 0.738 | 15 | 0.6 | 0.30 |
| -1.036 | -0.995 | 0.776 | 0.224 | 0.747 | 16 | 0.6 | 0.30 |
| -0.995 | -0.954 | 0.773 | 0.227 | 0.756 | 17 | 0.6 | 0.30 |
| -0.954 | -0.916 | 0.771 | 0.229 | 0.764 | 18 | 0.6 | 0.30 |
| -0.916 | -0.878 | 0.768 | 0.232 | 0.772 | 19 | 0.6 | 0.30 |
| -0.878 | -0.842 | 0.766 | 0.234 | 0.780 | 20 | 0.6 | 0.30 |
| -0.842 | -0.807 | 0.764 | 0.236 | 0.788 | 21 | 0.6 | 0.30 |
| -0.807 | -0.773 | 0.761 | 0.239 | 0.796 | 22 | 0.6 | 0.30 |
| -0.773 | -0.739 | 0.759 | 0.241 | 0.803 | 23 | 0.6 | 0.30 |
| -0.739 | -0.707 | 0.757 | 0.243 | 0.811 | 24 | 0.6 | 0.30 |
| -0.707 | -0.675 | 0.755 | 0.245 | 0.818 | 25 | 0.6 | 0.30 |
| -0.675 | -0.644 | 0.753 | 0.247 | 0.825 | 26 | 0.6 | 0.30 |
| -0.644 | -0.613 | 0.750 | 0.250 | 0.832 | 27 | 0.6 | 0.30 |
| -0.613 | -0.583 | 0.748 | 0.252 | 0.839 | 28 | 0.6 | 0.30 |
| -0.583 | -0.554 | 0.746 | 0.254 | 0.845 | 29 | 0.6 | 0.30 |
| -0.554 | -0.525 | 0.744 | 0.256 | 0.852 | 30 | 0.6 | 0.30 |
| -0.525 | -0.496 | 0.742 | 0.258 | 0.859 | 31 | 0.6 | 0.30 |
| -0.496 | -0.468 | 0.740 | 0.260 | 0.865 | 32 | 0.6 | 0.30 |
| -0.468 | -0.440 | 0.738 | 0.262 | 0.872 | 33 | 0.6 | 0.30 |
| -0.440 | -0.413 | 0.737 | 0.263 | 0.878 | 34 | 0.6 | 0.30 |
| -0.413 | -0.386 | 0.735 | 0.265 | 0.885 | 35 | 0.6 | 0.30 |
| -0.386 | -0.359 | 0.733 | 0.267 | 0.891 | 36 | 0.6 | 0.30 |
| -0.359 | -0.332 | 0.731 | 0.269 | 0.897 | 37 | 0.6 | 0.30 |
| -0.332 | -0.306 | 0.729 | 0.271 | 0.904 | 38 | 0.6 | 0.30 |
| -0.306 | -0.280 | 0.727 | 0.273 | 0.910 | 39 | 0.6 | 0.30 |
| -0.280 | -0.254 | 0.725 | 0.275 | 0.916 | 40 | 0.6 | 0.30 |
| -0.254 | -0.228 | 0.723 | 0.277 | 0.923 | 41 | 0.6 | 0.30 |
| -0.228 | -0.203 | 0.721 | 0.279 | 0.929 | 42 | 0.6 | 0.30 |
| -0.203 | -0.177 | 0.719 | 0.281 | 0.935 | 43 | 0.6 | 0.30 |
| -0.177 | -0.152 | 0.718 | 0.282 | 0.941 | 44 | 0.6 | 0.30 |
| -0.152 | -0.126 | 0.716 | 0.284 | 0.948 | 45 | 0.6 | 0.30 |
| -0.126 | -0.101 | 0.714 | 0.286 | 0.954 | 46 | 0.6 | 0.30 |
| -0.101 | -0.076 | 0.712 | 0.288 | 0.960 | 47 | 0.6 | 0.30 |
| -0.076 | -0.051 | 0.710 | 0.290 | 0.966 | 48 | 0.6 | 0.30 |
| -0.051 | -0.026 | 0.708 | 0.292 | 0.973 | 49 | 0.6 | 0.30 |
| -0.026 | -0.001 | 0.706 | 0.294 | 0.979 | 50 | 0.6 | 0.30 |
| -0.001 | 0.024 | 0.704 | 0.296 | 0.985 | 51 | 0.6 | 0.30 |
| 0.024 | 0.050 | 0.703 | 0.297 | 0.991 | 52 | 0.6 | 0.30 |
| 0.050 | 0.075 | 0.701 | 0.299 | 0.998 | 53 | 0.6 | 0.30 |
| 0.075 | 0.100 | 0.699 | 0.301 | 1.004 | 54 | 0.6 | 0.30 |
| 0.100 | 0.125 | 0.697 | 0.303 | 1.011 | 55 | 0.6 | 0.30 |
| 0.125 | 0.150 | 0.695 | 0.305 | 1.017 | 56 | 0.6 | 0.30 |
| 0.150 | 0.176 | 0.693 | 0.307 | 1.024 | 57 | 0.6 | 0.30 |
| 0.176 | 0.201 | 0.691 | 0.309 | 1.030 | 58 | 0.6 | 0.30 |
| 0.201 | 0.227 | 0.689 | 0.311 | 1.037 | 59 | 0.6 | 0.30 |
| 0.227 | 0.253 | 0.687 | 0.313 | 1.044 | 60 | 0.6 | 0.30 |
| 0.253 | 0.279 | 0.685 | 0.315 | 1.050 | 61 | 0.6 | 0.30 |
| 0.279 | 0.305 | 0.683 | 0.317 | 1.057 | 62 | 0.6 | 0.30 |
| 0.305 | 0.331 | 0.681 | 0.319 | 1.064 | 63 | 0.6 | 0.30 |
| 0.331 | 0.358 | 0.679 | 0.321 | 1.071 | 64 | 0.6 | 0.30 |
| 0.358 | 0.385 | 0.677 | 0.323 | 1.078 | 65 | 0.6 | 0.30 |
| 0.385 | 0.412 | 0.674 | 0.326 | 1.085 | 66 | 0.6 | 0.30 |
| 0.412 | 0.439 | 0.672 | 0.328 | 1.092 | 67 | 0.6 | 0.30 |
| 0.439 | 0.467 | 0.670 | 0.330 | 1.100 | 68 | 0.6 | 0.30 |
| 0.467 | 0.495 | 0.668 | 0.332 | 1.107 | 69 | 0.6 | 0.30 |
| 0.495 | 0.524 | 0.666 | 0.334 | 1.115 | 70 | 0.6 | 0.30 |
| 0.524 | 0.553 | 0.663 | 0.337 | 1.123 | 71 | 0.6 | 0.30 |
| 0.553 | 0.582 | 0.661 | 0.339 | 1.131 | 72 | 0.6 | 0.30 |
| 0.582 | 0.612 | 0.658 | 0.342 | 1.139 | 73 | 0.6 | 0.30 |
| 0.612 | 0.643 | 0.656 | 0.344 | 1.147 | 74 | 0.6 | 0.30 |
| 0.643 | 0.674 | 0.653 | 0.347 | 1.155 | 75 | 0.6 | 0.30 |
| 0.674 | 0.706 | 0.651 | 0.349 | 1.164 | 76 | 0.6 | 0.30 |
| 0.706 | 0.739 | 0.648 | 0.352 | 1.173 | 77 | 0.6 | 0.30 |
| 0.739 | 0.772 | 0.645 | 0.355 | 1.182 | 78 | 0.6 | 0.30 |
| 0.772 | 0.806 | 0.643 | 0.357 | 1.191 | 79 | 0.6 | 0.30 |
| 0.806 | 0.842 | 0.640 | 0.360 | 1.201 | 80 | 0.6 | 0.30 |
| 0.842 | 0.878 | 0.637 | 0.363 | 1.211 | 81 | 0.6 | 0.30 |
| 0.878 | 0.915 | 0.634 | 0.366 | 1.222 | 82 | 0.6 | 0.30 |
| 0.915 | 0.954 | 0.630 | 0.370 | 1.232 | 83 | 0.6 | 0.30 |
| 0.954 | 0.995 | 0.627 | 0.373 | 1.243 | 84 | 0.6 | 0.30 |
| 0.995 | 1.037 | 0.623 | 0.377 | 1.255 | 85 | 0.6 | 0.30 |
| 1.037 | 1.081 | 0.620 | 0.380 | 1.267 | 86 | 0.6 | 0.30 |
| 1.081 | 1.127 | 0.616 | 0.384 | 1.280 | 87 | 0.6 | 0.30 |
| 1.127 | 1.175 | 0.612 | 0.388 | 1.294 | 88 | 0.6 | 0.30 |
| 1.175 | 1.227 | 0.608 | 0.392 | 1.308 | 89 | 0.6 | 0.30 |
| 1.227 | 1.282 | 0.603 | 0.397 | 1.324 | 90 | 0.6 | 0.30 |
| 1.282 | 1.341 | 0.598 | 0.402 | 1.340 | 91 | 0.6 | 0.30 |
| 1.341 | 1.406 | 0.593 | 0.407 | 1.358 | 92 | 0.6 | 0.30 |
| 1.406 | 1.477 | 0.587 | 0.413 | 1.378 | 93 | 0.6 | 0.30 |
| 1.477 | 1.556 | 0.580 | 0.420 | 1.400 | 94 | 0.6 | 0.30 |
| 1.556 | 1.646 | 0.573 | 0.427 | 1.425 | 95 | 0.6 | 0.30 |
| 1.646 | 1.752 | 0.564 | 0.436 | 1.454 | 96 | 0.6 | 0.30 |
| 1.752 | 1.882 | 0.553 | 0.447 | 1.489 | 97 | 0.6 | 0.30 |
| 1.882 | 2.056 | 0.540 | 0.460 | 1.534 | 98 | 0.6 | 0.30 |
| 2.056 | 2.329 | 0.520 | 0.480 | 1.598 | 99 | 0.6 | 0.30 |
| 2.329 | Inf | 0.476 | 0.524 | 1.746 | 100 | 0.6 | 0.30 |
| -Inf | -2.302 | 0.952 | 0.048 | 0.159 | 1 | 0.7 | 0.30 |
| -2.302 | -2.036 | 0.934 | 0.066 | 0.221 | 2 | 0.7 | 0.30 |
| -2.036 | -1.867 | 0.923 | 0.077 | 0.257 | 3 | 0.7 | 0.30 |
| -1.867 | -1.739 | 0.914 | 0.086 | 0.286 | 4 | 0.7 | 0.30 |
| -1.739 | -1.636 | 0.907 | 0.093 | 0.310 | 5 | 0.7 | 0.30 |
| -1.636 | -1.547 | 0.901 | 0.099 | 0.332 | 6 | 0.7 | 0.30 |
| -1.547 | -1.470 | 0.895 | 0.105 | 0.351 | 7 | 0.7 | 0.30 |
| -1.470 | -1.400 | 0.889 | 0.111 | 0.370 | 8 | 0.7 | 0.30 |
| -1.400 | -1.337 | 0.884 | 0.116 | 0.387 | 9 | 0.7 | 0.30 |
| -1.337 | -1.279 | 0.879 | 0.121 | 0.404 | 10 | 0.7 | 0.30 |
| -1.279 | -1.224 | 0.874 | 0.126 | 0.420 | 11 | 0.7 | 0.30 |
| -1.224 | -1.174 | 0.869 | 0.131 | 0.435 | 12 | 0.7 | 0.30 |
| -1.174 | -1.126 | 0.865 | 0.135 | 0.450 | 13 | 0.7 | 0.30 |
| -1.126 | -1.080 | 0.861 | 0.139 | 0.464 | 14 | 0.7 | 0.30 |
| -1.080 | -1.037 | 0.856 | 0.144 | 0.478 | 15 | 0.7 | 0.30 |
| -1.037 | -0.995 | 0.852 | 0.148 | 0.492 | 16 | 0.7 | 0.30 |
| -0.995 | -0.955 | 0.848 | 0.152 | 0.506 | 17 | 0.7 | 0.30 |
| -0.955 | -0.917 | 0.844 | 0.156 | 0.519 | 18 | 0.7 | 0.30 |
| -0.917 | -0.880 | 0.840 | 0.160 | 0.532 | 19 | 0.7 | 0.30 |
| -0.880 | -0.844 | 0.836 | 0.164 | 0.545 | 20 | 0.7 | 0.30 |
| -0.844 | -0.809 | 0.833 | 0.167 | 0.558 | 21 | 0.7 | 0.30 |
| -0.809 | -0.775 | 0.829 | 0.171 | 0.570 | 22 | 0.7 | 0.30 |
| -0.775 | -0.742 | 0.825 | 0.175 | 0.583 | 23 | 0.7 | 0.30 |
| -0.742 | -0.710 | 0.821 | 0.179 | 0.595 | 24 | 0.7 | 0.30 |
| -0.710 | -0.678 | 0.818 | 0.182 | 0.608 | 25 | 0.7 | 0.30 |
| -0.678 | -0.647 | 0.814 | 0.186 | 0.620 | 26 | 0.7 | 0.30 |
| -0.647 | -0.617 | 0.810 | 0.190 | 0.632 | 27 | 0.7 | 0.30 |
| -0.617 | -0.587 | 0.807 | 0.193 | 0.644 | 28 | 0.7 | 0.30 |
| -0.587 | -0.558 | 0.803 | 0.197 | 0.656 | 29 | 0.7 | 0.30 |
| -0.558 | -0.529 | 0.799 | 0.201 | 0.668 | 30 | 0.7 | 0.30 |
| -0.529 | -0.501 | 0.796 | 0.204 | 0.681 | 31 | 0.7 | 0.30 |
| -0.501 | -0.473 | 0.792 | 0.208 | 0.693 | 32 | 0.7 | 0.30 |
| -0.473 | -0.445 | 0.789 | 0.211 | 0.705 | 33 | 0.7 | 0.30 |
| -0.445 | -0.418 | 0.785 | 0.215 | 0.717 | 34 | 0.7 | 0.30 |
| -0.418 | -0.391 | 0.781 | 0.219 | 0.729 | 35 | 0.7 | 0.30 |
| -0.391 | -0.364 | 0.778 | 0.222 | 0.741 | 36 | 0.7 | 0.30 |
| -0.364 | -0.337 | 0.774 | 0.226 | 0.753 | 37 | 0.7 | 0.30 |
| -0.337 | -0.311 | 0.770 | 0.230 | 0.765 | 38 | 0.7 | 0.30 |
| -0.311 | -0.285 | 0.767 | 0.233 | 0.777 | 39 | 0.7 | 0.30 |
| -0.285 | -0.259 | 0.763 | 0.237 | 0.789 | 40 | 0.7 | 0.30 |
| -0.259 | -0.233 | 0.759 | 0.241 | 0.802 | 41 | 0.7 | 0.30 |
| -0.233 | -0.207 | 0.756 | 0.244 | 0.814 | 42 | 0.7 | 0.30 |
| -0.207 | -0.182 | 0.752 | 0.248 | 0.826 | 43 | 0.7 | 0.30 |
| -0.182 | -0.157 | 0.748 | 0.252 | 0.839 | 44 | 0.7 | 0.30 |
| -0.157 | -0.131 | 0.745 | 0.255 | 0.851 | 45 | 0.7 | 0.30 |
| -0.131 | -0.106 | 0.741 | 0.259 | 0.864 | 46 | 0.7 | 0.30 |
| -0.106 | -0.081 | 0.737 | 0.263 | 0.877 | 47 | 0.7 | 0.30 |
| -0.081 | -0.056 | 0.733 | 0.267 | 0.889 | 48 | 0.7 | 0.30 |
| -0.056 | -0.031 | 0.729 | 0.271 | 0.902 | 49 | 0.7 | 0.30 |
| -0.031 | -0.005 | 0.725 | 0.275 | 0.915 | 50 | 0.7 | 0.30 |
| -0.005 | 0.020 | 0.721 | 0.279 | 0.928 | 51 | 0.7 | 0.30 |
| 0.020 | 0.045 | 0.717 | 0.283 | 0.942 | 52 | 0.7 | 0.30 |
| 0.045 | 0.070 | 0.713 | 0.287 | 0.955 | 53 | 0.7 | 0.30 |
| 0.070 | 0.095 | 0.709 | 0.291 | 0.969 | 54 | 0.7 | 0.30 |
| 0.095 | 0.121 | 0.705 | 0.295 | 0.982 | 55 | 0.7 | 0.30 |
| 0.121 | 0.146 | 0.701 | 0.299 | 0.996 | 56 | 0.7 | 0.30 |
| 0.146 | 0.171 | 0.697 | 0.303 | 1.010 | 57 | 0.7 | 0.30 |
| 0.171 | 0.197 | 0.693 | 0.307 | 1.024 | 58 | 0.7 | 0.30 |
| 0.197 | 0.223 | 0.688 | 0.312 | 1.039 | 59 | 0.7 | 0.30 |
| 0.223 | 0.249 | 0.684 | 0.316 | 1.053 | 60 | 0.7 | 0.30 |
| 0.249 | 0.275 | 0.680 | 0.320 | 1.068 | 61 | 0.7 | 0.30 |
| 0.275 | 0.301 | 0.675 | 0.325 | 1.083 | 62 | 0.7 | 0.30 |
| 0.301 | 0.328 | 0.671 | 0.329 | 1.098 | 63 | 0.7 | 0.30 |
| 0.328 | 0.354 | 0.666 | 0.334 | 1.113 | 64 | 0.7 | 0.30 |
| 0.354 | 0.381 | 0.661 | 0.339 | 1.129 | 65 | 0.7 | 0.30 |
| 0.381 | 0.409 | 0.657 | 0.343 | 1.145 | 66 | 0.7 | 0.30 |
| 0.409 | 0.436 | 0.652 | 0.348 | 1.161 | 67 | 0.7 | 0.30 |
| 0.436 | 0.464 | 0.647 | 0.353 | 1.178 | 68 | 0.7 | 0.30 |
| 0.464 | 0.493 | 0.642 | 0.358 | 1.195 | 69 | 0.7 | 0.30 |
| 0.493 | 0.521 | 0.636 | 0.364 | 1.212 | 70 | 0.7 | 0.30 |
| 0.521 | 0.551 | 0.631 | 0.369 | 1.229 | 71 | 0.7 | 0.30 |
| 0.551 | 0.580 | 0.626 | 0.374 | 1.247 | 72 | 0.7 | 0.30 |
| 0.580 | 0.611 | 0.620 | 0.380 | 1.266 | 73 | 0.7 | 0.30 |
| 0.611 | 0.641 | 0.615 | 0.385 | 1.284 | 74 | 0.7 | 0.30 |
| 0.641 | 0.673 | 0.609 | 0.391 | 1.304 | 75 | 0.7 | 0.30 |
| 0.673 | 0.705 | 0.603 | 0.397 | 1.323 | 76 | 0.7 | 0.30 |
| 0.705 | 0.738 | 0.597 | 0.403 | 1.344 | 77 | 0.7 | 0.30 |
| 0.738 | 0.771 | 0.591 | 0.409 | 1.365 | 78 | 0.7 | 0.30 |
| 0.771 | 0.806 | 0.584 | 0.416 | 1.386 | 79 | 0.7 | 0.30 |
| 0.806 | 0.842 | 0.577 | 0.423 | 1.409 | 80 | 0.7 | 0.30 |
| 0.842 | 0.878 | 0.570 | 0.430 | 1.432 | 81 | 0.7 | 0.30 |
| 0.878 | 0.916 | 0.563 | 0.437 | 1.456 | 82 | 0.7 | 0.30 |
| 0.916 | 0.955 | 0.556 | 0.444 | 1.480 | 83 | 0.7 | 0.30 |
| 0.955 | 0.996 | 0.548 | 0.452 | 1.506 | 84 | 0.7 | 0.30 |
| 0.996 | 1.038 | 0.540 | 0.460 | 1.533 | 85 | 0.7 | 0.30 |
| 1.038 | 1.083 | 0.532 | 0.468 | 1.561 | 86 | 0.7 | 0.30 |
| 1.083 | 1.129 | 0.523 | 0.477 | 1.591 | 87 | 0.7 | 0.30 |
| 1.129 | 1.179 | 0.513 | 0.487 | 1.622 | 88 | 0.7 | 0.30 |
| 1.179 | 1.231 | 0.503 | 0.497 | 1.655 | 89 | 0.7 | 0.30 |
| 1.231 | 1.287 | 0.493 | 0.507 | 1.690 | 90 | 0.7 | 0.30 |
| 1.287 | 1.346 | 0.482 | 0.518 | 1.728 | 91 | 0.7 | 0.30 |
| 1.346 | 1.412 | 0.469 | 0.531 | 1.769 | 92 | 0.7 | 0.30 |
| 1.412 | 1.483 | 0.456 | 0.544 | 1.813 | 93 | 0.7 | 0.30 |
| 1.483 | 1.563 | 0.441 | 0.559 | 1.862 | 94 | 0.7 | 0.30 |
| 1.563 | 1.654 | 0.425 | 0.575 | 1.916 | 95 | 0.7 | 0.30 |
| 1.654 | 1.762 | 0.406 | 0.594 | 1.979 | 96 | 0.7 | 0.30 |
| 1.762 | 1.893 | 0.384 | 0.616 | 2.053 | 97 | 0.7 | 0.30 |
| 1.893 | 2.068 | 0.357 | 0.643 | 2.145 | 98 | 0.7 | 0.30 |
| 2.068 | 2.344 | 0.319 | 0.681 | 2.271 | 99 | 0.7 | 0.30 |
| 2.344 | Inf | 0.244 | 0.756 | 2.520 | 100 | 0.7 | 0.30 |
| -Inf | -2.239 | 0.989 | 0.011 | 0.036 | 1 | 0.8 | 0.30 |
| -2.239 | -1.989 | 0.982 | 0.018 | 0.061 | 2 | 0.8 | 0.30 |
| -1.989 | -1.829 | 0.976 | 0.024 | 0.080 | 3 | 0.8 | 0.30 |
| -1.829 | -1.709 | 0.971 | 0.029 | 0.095 | 4 | 0.8 | 0.30 |
| -1.709 | -1.610 | 0.967 | 0.033 | 0.110 | 5 | 0.8 | 0.30 |
| -1.610 | -1.526 | 0.963 | 0.037 | 0.124 | 6 | 0.8 | 0.30 |
| -1.526 | -1.452 | 0.959 | 0.041 | 0.137 | 7 | 0.8 | 0.30 |
| -1.452 | -1.386 | 0.955 | 0.045 | 0.150 | 8 | 0.8 | 0.30 |
| -1.386 | -1.326 | 0.951 | 0.049 | 0.163 | 9 | 0.8 | 0.30 |
| -1.326 | -1.270 | 0.947 | 0.053 | 0.176 | 10 | 0.8 | 0.30 |
| -1.270 | -1.218 | 0.944 | 0.056 | 0.188 | 11 | 0.8 | 0.30 |
| -1.218 | -1.169 | 0.940 | 0.060 | 0.201 | 12 | 0.8 | 0.30 |
| -1.169 | -1.123 | 0.936 | 0.064 | 0.213 | 13 | 0.8 | 0.30 |
| -1.123 | -1.079 | 0.932 | 0.068 | 0.226 | 14 | 0.8 | 0.30 |
| -1.079 | -1.037 | 0.929 | 0.071 | 0.238 | 15 | 0.8 | 0.30 |
| -1.037 | -0.997 | 0.925 | 0.075 | 0.251 | 16 | 0.8 | 0.30 |
| -0.997 | -0.959 | 0.921 | 0.079 | 0.263 | 17 | 0.8 | 0.30 |
| -0.959 | -0.921 | 0.917 | 0.083 | 0.276 | 18 | 0.8 | 0.30 |
| -0.921 | -0.885 | 0.913 | 0.087 | 0.289 | 19 | 0.8 | 0.30 |
| -0.885 | -0.850 | 0.909 | 0.091 | 0.302 | 20 | 0.8 | 0.30 |
| -0.850 | -0.817 | 0.906 | 0.094 | 0.315 | 21 | 0.8 | 0.30 |
| -0.817 | -0.783 | 0.902 | 0.098 | 0.328 | 22 | 0.8 | 0.30 |
| -0.783 | -0.751 | 0.898 | 0.102 | 0.341 | 23 | 0.8 | 0.30 |
| -0.751 | -0.720 | 0.894 | 0.106 | 0.355 | 24 | 0.8 | 0.30 |
| -0.720 | -0.689 | 0.889 | 0.111 | 0.368 | 25 | 0.8 | 0.30 |
| -0.689 | -0.659 | 0.885 | 0.115 | 0.382 | 26 | 0.8 | 0.30 |
| -0.659 | -0.629 | 0.881 | 0.119 | 0.396 | 27 | 0.8 | 0.30 |
| -0.629 | -0.600 | 0.877 | 0.123 | 0.410 | 28 | 0.8 | 0.30 |
| -0.600 | -0.571 | 0.873 | 0.127 | 0.425 | 29 | 0.8 | 0.30 |
| -0.571 | -0.542 | 0.868 | 0.132 | 0.439 | 30 | 0.8 | 0.30 |
| -0.542 | -0.514 | 0.864 | 0.136 | 0.454 | 31 | 0.8 | 0.30 |
| -0.514 | -0.487 | 0.859 | 0.141 | 0.469 | 32 | 0.8 | 0.30 |
| -0.487 | -0.460 | 0.855 | 0.145 | 0.484 | 33 | 0.8 | 0.30 |
| -0.460 | -0.432 | 0.850 | 0.150 | 0.500 | 34 | 0.8 | 0.30 |
| -0.432 | -0.406 | 0.845 | 0.155 | 0.515 | 35 | 0.8 | 0.30 |
| -0.406 | -0.379 | 0.841 | 0.159 | 0.531 | 36 | 0.8 | 0.30 |
| -0.379 | -0.353 | 0.836 | 0.164 | 0.548 | 37 | 0.8 | 0.30 |
| -0.353 | -0.327 | 0.831 | 0.169 | 0.564 | 38 | 0.8 | 0.30 |
| -0.327 | -0.301 | 0.826 | 0.174 | 0.581 | 39 | 0.8 | 0.30 |
| -0.301 | -0.275 | 0.821 | 0.179 | 0.598 | 40 | 0.8 | 0.30 |
| -0.275 | -0.249 | 0.815 | 0.185 | 0.615 | 41 | 0.8 | 0.30 |
| -0.249 | -0.224 | 0.810 | 0.190 | 0.633 | 42 | 0.8 | 0.30 |
| -0.224 | -0.198 | 0.805 | 0.195 | 0.651 | 43 | 0.8 | 0.30 |
| -0.198 | -0.173 | 0.799 | 0.201 | 0.669 | 44 | 0.8 | 0.30 |
| -0.173 | -0.147 | 0.794 | 0.206 | 0.687 | 45 | 0.8 | 0.30 |
| -0.147 | -0.122 | 0.788 | 0.212 | 0.706 | 46 | 0.8 | 0.30 |
| -0.122 | -0.097 | 0.782 | 0.218 | 0.726 | 47 | 0.8 | 0.30 |
| -0.097 | -0.072 | 0.776 | 0.224 | 0.745 | 48 | 0.8 | 0.30 |
| -0.072 | -0.046 | 0.770 | 0.230 | 0.765 | 49 | 0.8 | 0.30 |
| -0.046 | -0.021 | 0.764 | 0.236 | 0.786 | 50 | 0.8 | 0.30 |
| -0.021 | 0.004 | 0.758 | 0.242 | 0.806 | 51 | 0.8 | 0.30 |
| 0.004 | 0.030 | 0.752 | 0.248 | 0.828 | 52 | 0.8 | 0.30 |
| 0.030 | 0.055 | 0.745 | 0.255 | 0.849 | 53 | 0.8 | 0.30 |
| 0.055 | 0.080 | 0.739 | 0.261 | 0.871 | 54 | 0.8 | 0.30 |
| 0.080 | 0.106 | 0.732 | 0.268 | 0.894 | 55 | 0.8 | 0.30 |
| 0.106 | 0.132 | 0.725 | 0.275 | 0.917 | 56 | 0.8 | 0.30 |
| 0.132 | 0.158 | 0.718 | 0.282 | 0.940 | 57 | 0.8 | 0.30 |
| 0.158 | 0.184 | 0.711 | 0.289 | 0.964 | 58 | 0.8 | 0.30 |
| 0.184 | 0.210 | 0.703 | 0.297 | 0.988 | 59 | 0.8 | 0.30 |
| 0.210 | 0.236 | 0.696 | 0.304 | 1.013 | 60 | 0.8 | 0.30 |
| 0.236 | 0.263 | 0.688 | 0.312 | 1.039 | 61 | 0.8 | 0.30 |
| 0.263 | 0.290 | 0.681 | 0.319 | 1.065 | 62 | 0.8 | 0.30 |
| 0.290 | 0.317 | 0.673 | 0.327 | 1.092 | 63 | 0.8 | 0.30 |
| 0.317 | 0.344 | 0.664 | 0.336 | 1.119 | 64 | 0.8 | 0.30 |
| 0.344 | 0.372 | 0.656 | 0.344 | 1.147 | 65 | 0.8 | 0.30 |
| 0.372 | 0.400 | 0.647 | 0.353 | 1.175 | 66 | 0.8 | 0.30 |
| 0.400 | 0.428 | 0.639 | 0.361 | 1.204 | 67 | 0.8 | 0.30 |
| 0.428 | 0.457 | 0.630 | 0.370 | 1.234 | 68 | 0.8 | 0.30 |
| 0.457 | 0.486 | 0.621 | 0.379 | 1.265 | 69 | 0.8 | 0.30 |
| 0.486 | 0.515 | 0.611 | 0.389 | 1.296 | 70 | 0.8 | 0.30 |
| 0.515 | 0.545 | 0.602 | 0.398 | 1.328 | 71 | 0.8 | 0.30 |
| 0.545 | 0.576 | 0.592 | 0.408 | 1.361 | 72 | 0.8 | 0.30 |
| 0.576 | 0.607 | 0.582 | 0.418 | 1.395 | 73 | 0.8 | 0.30 |
| 0.607 | 0.639 | 0.571 | 0.429 | 1.430 | 74 | 0.8 | 0.30 |
| 0.639 | 0.671 | 0.560 | 0.440 | 1.465 | 75 | 0.8 | 0.30 |
| 0.671 | 0.704 | 0.550 | 0.450 | 1.502 | 76 | 0.8 | 0.30 |
| 0.704 | 0.738 | 0.538 | 0.462 | 1.539 | 77 | 0.8 | 0.30 |
| 0.738 | 0.773 | 0.527 | 0.473 | 1.578 | 78 | 0.8 | 0.30 |
| 0.773 | 0.808 | 0.515 | 0.485 | 1.617 | 79 | 0.8 | 0.30 |
| 0.808 | 0.845 | 0.503 | 0.497 | 1.658 | 80 | 0.8 | 0.30 |
| 0.845 | 0.883 | 0.490 | 0.510 | 1.700 | 81 | 0.8 | 0.30 |
| 0.883 | 0.922 | 0.477 | 0.523 | 1.744 | 82 | 0.8 | 0.30 |
| 0.922 | 0.962 | 0.463 | 0.537 | 1.789 | 83 | 0.8 | 0.30 |
| 0.962 | 1.005 | 0.450 | 0.550 | 1.835 | 84 | 0.8 | 0.30 |
| 1.005 | 1.048 | 0.435 | 0.565 | 1.883 | 85 | 0.8 | 0.30 |
| 1.048 | 1.094 | 0.420 | 0.580 | 1.932 | 86 | 0.8 | 0.30 |
| 1.094 | 1.142 | 0.405 | 0.595 | 1.984 | 87 | 0.8 | 0.30 |
| 1.142 | 1.193 | 0.389 | 0.611 | 2.037 | 88 | 0.8 | 0.30 |
| 1.193 | 1.247 | 0.372 | 0.628 | 2.093 | 89 | 0.8 | 0.30 |
| 1.247 | 1.304 | 0.355 | 0.645 | 2.151 | 90 | 0.8 | 0.30 |
| 1.304 | 1.366 | 0.337 | 0.663 | 2.212 | 91 | 0.8 | 0.30 |
| 1.366 | 1.433 | 0.317 | 0.683 | 2.275 | 92 | 0.8 | 0.30 |
| 1.433 | 1.506 | 0.297 | 0.703 | 2.343 | 93 | 0.8 | 0.30 |
| 1.506 | 1.588 | 0.276 | 0.724 | 2.414 | 94 | 0.8 | 0.30 |
| 1.588 | 1.681 | 0.253 | 0.747 | 2.491 | 95 | 0.8 | 0.30 |
| 1.681 | 1.790 | 0.228 | 0.772 | 2.573 | 96 | 0.8 | 0.30 |
| 1.790 | 1.923 | 0.201 | 0.799 | 2.664 | 97 | 0.8 | 0.30 |
| 1.923 | 2.099 | 0.170 | 0.830 | 2.768 | 98 | 0.8 | 0.30 |
| 2.099 | 2.373 | 0.133 | 0.867 | 2.892 | 99 | 0.8 | 0.30 |
| 2.373 | Inf | 0.077 | 0.923 | 3.075 | 100 | 0.8 | 0.30 |
| -Inf | -2.103 | 0.999 | 0.001 | 0.003 | 1 | 0.9 | 0.30 |
| -2.103 | -1.882 | 0.998 | 0.002 | 0.007 | 2 | 0.9 | 0.30 |
| -1.882 | -1.741 | 0.997 | 0.003 | 0.010 | 3 | 0.9 | 0.30 |
| -1.741 | -1.634 | 0.996 | 0.004 | 0.014 | 4 | 0.9 | 0.30 |
| -1.634 | -1.547 | 0.995 | 0.005 | 0.017 | 5 | 0.9 | 0.30 |
| -1.547 | -1.472 | 0.994 | 0.006 | 0.021 | 6 | 0.9 | 0.30 |
| -1.472 | -1.406 | 0.993 | 0.007 | 0.025 | 7 | 0.9 | 0.30 |
| -1.406 | -1.347 | 0.991 | 0.009 | 0.029 | 8 | 0.9 | 0.30 |
| -1.347 | -1.292 | 0.990 | 0.010 | 0.033 | 9 | 0.9 | 0.30 |
| -1.292 | -1.242 | 0.989 | 0.011 | 0.037 | 10 | 0.9 | 0.30 |
| -1.242 | -1.196 | 0.988 | 0.012 | 0.041 | 11 | 0.9 | 0.30 |
| -1.196 | -1.152 | 0.986 | 0.014 | 0.046 | 12 | 0.9 | 0.30 |
| -1.152 | -1.110 | 0.985 | 0.015 | 0.051 | 13 | 0.9 | 0.30 |
| -1.110 | -1.070 | 0.983 | 0.017 | 0.056 | 14 | 0.9 | 0.30 |
| -1.070 | -1.032 | 0.982 | 0.018 | 0.061 | 15 | 0.9 | 0.30 |
| -1.032 | -0.996 | 0.980 | 0.020 | 0.067 | 16 | 0.9 | 0.30 |
| -0.996 | -0.961 | 0.978 | 0.022 | 0.072 | 17 | 0.9 | 0.30 |
| -0.961 | -0.927 | 0.977 | 0.023 | 0.078 | 18 | 0.9 | 0.30 |
| -0.927 | -0.894 | 0.975 | 0.025 | 0.085 | 19 | 0.9 | 0.30 |
| -0.894 | -0.862 | 0.973 | 0.027 | 0.091 | 20 | 0.9 | 0.30 |
| -0.862 | -0.831 | 0.971 | 0.029 | 0.098 | 21 | 0.9 | 0.30 |
| -0.831 | -0.800 | 0.968 | 0.032 | 0.105 | 22 | 0.9 | 0.30 |
| -0.800 | -0.770 | 0.966 | 0.034 | 0.113 | 23 | 0.9 | 0.30 |
| -0.770 | -0.741 | 0.964 | 0.036 | 0.120 | 24 | 0.9 | 0.30 |
| -0.741 | -0.712 | 0.961 | 0.039 | 0.129 | 25 | 0.9 | 0.30 |
| -0.712 | -0.684 | 0.959 | 0.041 | 0.137 | 26 | 0.9 | 0.30 |
| -0.684 | -0.656 | 0.956 | 0.044 | 0.146 | 27 | 0.9 | 0.30 |
| -0.656 | -0.629 | 0.953 | 0.047 | 0.155 | 28 | 0.9 | 0.30 |
| -0.629 | -0.602 | 0.950 | 0.050 | 0.165 | 29 | 0.9 | 0.30 |
| -0.602 | -0.575 | 0.947 | 0.053 | 0.175 | 30 | 0.9 | 0.30 |
| -0.575 | -0.549 | 0.944 | 0.056 | 0.186 | 31 | 0.9 | 0.30 |
| -0.549 | -0.522 | 0.941 | 0.059 | 0.197 | 32 | 0.9 | 0.30 |
| -0.522 | -0.496 | 0.937 | 0.063 | 0.209 | 33 | 0.9 | 0.30 |
| -0.496 | -0.471 | 0.934 | 0.066 | 0.221 | 34 | 0.9 | 0.30 |
| -0.471 | -0.445 | 0.930 | 0.070 | 0.234 | 35 | 0.9 | 0.30 |
| -0.445 | -0.420 | 0.926 | 0.074 | 0.248 | 36 | 0.9 | 0.30 |
| -0.420 | -0.394 | 0.921 | 0.079 | 0.262 | 37 | 0.9 | 0.30 |
| -0.394 | -0.369 | 0.917 | 0.083 | 0.277 | 38 | 0.9 | 0.30 |
| -0.369 | -0.344 | 0.912 | 0.088 | 0.292 | 39 | 0.9 | 0.30 |
| -0.344 | -0.318 | 0.908 | 0.092 | 0.308 | 40 | 0.9 | 0.30 |
| -0.318 | -0.293 | 0.902 | 0.098 | 0.325 | 41 | 0.9 | 0.30 |
| -0.293 | -0.268 | 0.897 | 0.103 | 0.343 | 42 | 0.9 | 0.30 |
| -0.268 | -0.243 | 0.892 | 0.108 | 0.362 | 43 | 0.9 | 0.30 |
| -0.243 | -0.218 | 0.886 | 0.114 | 0.381 | 44 | 0.9 | 0.30 |
| -0.218 | -0.193 | 0.880 | 0.120 | 0.402 | 45 | 0.9 | 0.30 |
| -0.193 | -0.168 | 0.873 | 0.127 | 0.423 | 46 | 0.9 | 0.30 |
| -0.168 | -0.142 | 0.866 | 0.134 | 0.445 | 47 | 0.9 | 0.30 |
| -0.142 | -0.117 | 0.859 | 0.141 | 0.469 | 48 | 0.9 | 0.30 |
| -0.117 | -0.091 | 0.852 | 0.148 | 0.494 | 49 | 0.9 | 0.30 |
| -0.091 | -0.066 | 0.844 | 0.156 | 0.520 | 50 | 0.9 | 0.30 |
| -0.066 | -0.040 | 0.836 | 0.164 | 0.547 | 51 | 0.9 | 0.30 |
| -0.040 | -0.014 | 0.827 | 0.173 | 0.575 | 52 | 0.9 | 0.30 |
| -0.014 | 0.012 | 0.819 | 0.181 | 0.605 | 53 | 0.9 | 0.30 |
| 0.012 | 0.038 | 0.809 | 0.191 | 0.636 | 54 | 0.9 | 0.30 |
| 0.038 | 0.065 | 0.799 | 0.201 | 0.669 | 55 | 0.9 | 0.30 |
| 0.065 | 0.092 | 0.789 | 0.211 | 0.703 | 56 | 0.9 | 0.30 |
| 0.092 | 0.119 | 0.778 | 0.222 | 0.739 | 57 | 0.9 | 0.30 |
| 0.119 | 0.146 | 0.767 | 0.233 | 0.776 | 58 | 0.9 | 0.30 |
| 0.146 | 0.174 | 0.755 | 0.245 | 0.815 | 59 | 0.9 | 0.30 |
| 0.174 | 0.202 | 0.743 | 0.257 | 0.856 | 60 | 0.9 | 0.30 |
| 0.202 | 0.230 | 0.730 | 0.270 | 0.899 | 61 | 0.9 | 0.30 |
| 0.230 | 0.258 | 0.717 | 0.283 | 0.944 | 62 | 0.9 | 0.30 |
| 0.258 | 0.287 | 0.703 | 0.297 | 0.990 | 63 | 0.9 | 0.30 |
| 0.287 | 0.317 | 0.688 | 0.312 | 1.039 | 64 | 0.9 | 0.30 |
| 0.317 | 0.347 | 0.673 | 0.327 | 1.089 | 65 | 0.9 | 0.30 |
| 0.347 | 0.377 | 0.657 | 0.343 | 1.142 | 66 | 0.9 | 0.30 |
| 0.377 | 0.408 | 0.641 | 0.359 | 1.197 | 67 | 0.9 | 0.30 |
| 0.408 | 0.439 | 0.624 | 0.376 | 1.253 | 68 | 0.9 | 0.30 |
| 0.439 | 0.471 | 0.606 | 0.394 | 1.312 | 69 | 0.9 | 0.30 |
| 0.471 | 0.503 | 0.588 | 0.412 | 1.372 | 70 | 0.9 | 0.30 |
| 0.503 | 0.536 | 0.570 | 0.430 | 1.435 | 71 | 0.9 | 0.30 |
| 0.536 | 0.570 | 0.550 | 0.450 | 1.499 | 72 | 0.9 | 0.30 |
| 0.570 | 0.604 | 0.530 | 0.470 | 1.566 | 73 | 0.9 | 0.30 |
| 0.604 | 0.639 | 0.510 | 0.490 | 1.634 | 74 | 0.9 | 0.30 |
| 0.639 | 0.675 | 0.489 | 0.511 | 1.703 | 75 | 0.9 | 0.30 |
| 0.675 | 0.712 | 0.468 | 0.532 | 1.774 | 76 | 0.9 | 0.30 |
| 0.712 | 0.749 | 0.446 | 0.554 | 1.846 | 77 | 0.9 | 0.30 |
| 0.749 | 0.787 | 0.424 | 0.576 | 1.919 | 78 | 0.9 | 0.30 |
| 0.787 | 0.827 | 0.402 | 0.598 | 1.993 | 79 | 0.9 | 0.30 |
| 0.827 | 0.867 | 0.380 | 0.620 | 2.068 | 80 | 0.9 | 0.30 |
| 0.867 | 0.909 | 0.357 | 0.643 | 2.143 | 81 | 0.9 | 0.30 |
| 0.909 | 0.952 | 0.335 | 0.665 | 2.218 | 82 | 0.9 | 0.30 |
| 0.952 | 0.996 | 0.312 | 0.688 | 2.292 | 83 | 0.9 | 0.30 |
| 0.996 | 1.042 | 0.290 | 0.710 | 2.367 | 84 | 0.9 | 0.30 |
| 1.042 | 1.089 | 0.268 | 0.732 | 2.440 | 85 | 0.9 | 0.30 |
| 1.089 | 1.138 | 0.246 | 0.754 | 2.513 | 86 | 0.9 | 0.30 |
| 1.138 | 1.190 | 0.225 | 0.775 | 2.584 | 87 | 0.9 | 0.30 |
| 1.190 | 1.243 | 0.204 | 0.796 | 2.653 | 88 | 0.9 | 0.30 |
| 1.243 | 1.300 | 0.184 | 0.816 | 2.720 | 89 | 0.9 | 0.30 |
| 1.300 | 1.360 | 0.164 | 0.836 | 2.786 | 90 | 0.9 | 0.30 |
| 1.360 | 1.423 | 0.145 | 0.855 | 2.849 | 91 | 0.9 | 0.30 |
| 1.423 | 1.491 | 0.127 | 0.873 | 2.910 | 92 | 0.9 | 0.30 |
| 1.491 | 1.565 | 0.110 | 0.890 | 2.968 | 93 | 0.9 | 0.30 |
| 1.565 | 1.647 | 0.093 | 0.907 | 3.023 | 94 | 0.9 | 0.30 |
| 1.647 | 1.739 | 0.077 | 0.923 | 3.076 | 95 | 0.9 | 0.30 |
| 1.739 | 1.844 | 0.062 | 0.938 | 3.125 | 96 | 0.9 | 0.30 |
| 1.844 | 1.972 | 0.048 | 0.952 | 3.173 | 97 | 0.9 | 0.30 |
| 1.972 | 2.137 | 0.035 | 0.965 | 3.217 | 98 | 0.9 | 0.30 |
| 2.137 | 2.390 | 0.022 | 0.978 | 3.259 | 99 | 0.9 | 0.30 |
| 2.390 | Inf | 0.009 | 0.991 | 3.303 | 100 | 0.9 | 0.30 |
| -Inf | -2.326 | 0.724 | 0.276 | 0.551 | 1 | 0.6 | 0.50 |
| -2.326 | -2.054 | 0.688 | 0.312 | 0.623 | 2 | 0.6 | 0.50 |
| -2.054 | -1.881 | 0.671 | 0.329 | 0.657 | 3 | 0.6 | 0.50 |
| -1.881 | -1.751 | 0.659 | 0.341 | 0.682 | 4 | 0.6 | 0.50 |
| -1.751 | -1.645 | 0.650 | 0.350 | 0.701 | 5 | 0.6 | 0.50 |
| -1.645 | -1.555 | 0.642 | 0.358 | 0.717 | 6 | 0.6 | 0.50 |
| -1.555 | -1.476 | 0.634 | 0.366 | 0.731 | 7 | 0.6 | 0.50 |
| -1.476 | -1.405 | 0.628 | 0.372 | 0.744 | 8 | 0.6 | 0.50 |
| -1.405 | -1.341 | 0.622 | 0.378 | 0.755 | 9 | 0.6 | 0.50 |
| -1.341 | -1.282 | 0.617 | 0.383 | 0.766 | 10 | 0.6 | 0.50 |
| -1.282 | -1.227 | 0.612 | 0.388 | 0.776 | 11 | 0.6 | 0.50 |
| -1.227 | -1.175 | 0.608 | 0.392 | 0.785 | 12 | 0.6 | 0.50 |
| -1.175 | -1.127 | 0.603 | 0.397 | 0.794 | 13 | 0.6 | 0.50 |
| -1.127 | -1.080 | 0.599 | 0.401 | 0.802 | 14 | 0.6 | 0.50 |
| -1.080 | -1.037 | 0.595 | 0.405 | 0.810 | 15 | 0.6 | 0.50 |
| -1.037 | -0.995 | 0.591 | 0.409 | 0.817 | 16 | 0.6 | 0.50 |
| -0.995 | -0.954 | 0.588 | 0.412 | 0.825 | 17 | 0.6 | 0.50 |
| -0.954 | -0.916 | 0.584 | 0.416 | 0.831 | 18 | 0.6 | 0.50 |
| -0.916 | -0.878 | 0.581 | 0.419 | 0.838 | 19 | 0.6 | 0.50 |
| -0.878 | -0.842 | 0.578 | 0.422 | 0.845 | 20 | 0.6 | 0.50 |
| -0.842 | -0.807 | 0.574 | 0.426 | 0.851 | 21 | 0.6 | 0.50 |
| -0.807 | -0.772 | 0.571 | 0.429 | 0.857 | 22 | 0.6 | 0.50 |
| -0.772 | -0.739 | 0.568 | 0.432 | 0.863 | 23 | 0.6 | 0.50 |
| -0.739 | -0.706 | 0.565 | 0.435 | 0.869 | 24 | 0.6 | 0.50 |
| -0.706 | -0.675 | 0.563 | 0.437 | 0.875 | 25 | 0.6 | 0.50 |
| -0.675 | -0.643 | 0.560 | 0.440 | 0.881 | 26 | 0.6 | 0.50 |
| -0.643 | -0.613 | 0.557 | 0.443 | 0.886 | 27 | 0.6 | 0.50 |
| -0.613 | -0.583 | 0.554 | 0.446 | 0.892 | 28 | 0.6 | 0.50 |
| -0.583 | -0.554 | 0.552 | 0.448 | 0.897 | 29 | 0.6 | 0.50 |
| -0.554 | -0.525 | 0.549 | 0.451 | 0.902 | 30 | 0.6 | 0.50 |
| -0.525 | -0.496 | 0.546 | 0.454 | 0.907 | 31 | 0.6 | 0.50 |
| -0.496 | -0.468 | 0.544 | 0.456 | 0.913 | 32 | 0.6 | 0.50 |
| -0.468 | -0.440 | 0.541 | 0.459 | 0.918 | 33 | 0.6 | 0.50 |
| -0.440 | -0.413 | 0.539 | 0.461 | 0.923 | 34 | 0.6 | 0.50 |
| -0.413 | -0.385 | 0.536 | 0.464 | 0.928 | 35 | 0.6 | 0.50 |
| -0.385 | -0.359 | 0.534 | 0.466 | 0.932 | 36 | 0.6 | 0.50 |
| -0.359 | -0.332 | 0.531 | 0.469 | 0.937 | 37 | 0.6 | 0.50 |
| -0.332 | -0.306 | 0.529 | 0.471 | 0.942 | 38 | 0.6 | 0.50 |
| -0.306 | -0.279 | 0.527 | 0.473 | 0.947 | 39 | 0.6 | 0.50 |
| -0.279 | -0.253 | 0.524 | 0.476 | 0.952 | 40 | 0.6 | 0.50 |
| -0.253 | -0.228 | 0.522 | 0.478 | 0.956 | 41 | 0.6 | 0.50 |
| -0.228 | -0.202 | 0.520 | 0.480 | 0.961 | 42 | 0.6 | 0.50 |
| -0.202 | -0.176 | 0.517 | 0.483 | 0.966 | 43 | 0.6 | 0.50 |
| -0.176 | -0.151 | 0.515 | 0.485 | 0.970 | 44 | 0.6 | 0.50 |
| -0.151 | -0.126 | 0.513 | 0.487 | 0.975 | 45 | 0.6 | 0.50 |
| -0.126 | -0.100 | 0.510 | 0.490 | 0.979 | 46 | 0.6 | 0.50 |
| -0.100 | -0.075 | 0.508 | 0.492 | 0.984 | 47 | 0.6 | 0.50 |
| -0.075 | -0.050 | 0.506 | 0.494 | 0.989 | 48 | 0.6 | 0.50 |
| -0.050 | -0.025 | 0.503 | 0.497 | 0.993 | 49 | 0.6 | 0.50 |
| -0.025 | 0.000 | 0.501 | 0.499 | 0.998 | 50 | 0.6 | 0.50 |
| 0.000 | 0.025 | 0.499 | 0.501 | 1.002 | 51 | 0.6 | 0.50 |
| 0.025 | 0.050 | 0.497 | 0.503 | 1.007 | 52 | 0.6 | 0.50 |
| 0.050 | 0.075 | 0.494 | 0.506 | 1.011 | 53 | 0.6 | 0.50 |
| 0.075 | 0.100 | 0.492 | 0.508 | 1.016 | 54 | 0.6 | 0.50 |
| 0.100 | 0.126 | 0.490 | 0.510 | 1.021 | 55 | 0.6 | 0.50 |
| 0.126 | 0.151 | 0.487 | 0.513 | 1.025 | 56 | 0.6 | 0.50 |
| 0.151 | 0.176 | 0.485 | 0.515 | 1.030 | 57 | 0.6 | 0.50 |
| 0.176 | 0.202 | 0.483 | 0.517 | 1.034 | 58 | 0.6 | 0.50 |
| 0.202 | 0.228 | 0.480 | 0.520 | 1.039 | 59 | 0.6 | 0.50 |
| 0.228 | 0.253 | 0.478 | 0.522 | 1.044 | 60 | 0.6 | 0.50 |
| 0.253 | 0.279 | 0.476 | 0.524 | 1.048 | 61 | 0.6 | 0.50 |
| 0.279 | 0.306 | 0.473 | 0.527 | 1.053 | 62 | 0.6 | 0.50 |
| 0.306 | 0.332 | 0.471 | 0.529 | 1.058 | 63 | 0.6 | 0.50 |
| 0.332 | 0.359 | 0.469 | 0.531 | 1.063 | 64 | 0.6 | 0.50 |
| 0.359 | 0.385 | 0.466 | 0.534 | 1.068 | 65 | 0.6 | 0.50 |
| 0.385 | 0.413 | 0.464 | 0.536 | 1.072 | 66 | 0.6 | 0.50 |
| 0.413 | 0.440 | 0.461 | 0.539 | 1.077 | 67 | 0.6 | 0.50 |
| 0.440 | 0.468 | 0.459 | 0.541 | 1.082 | 68 | 0.6 | 0.50 |
| 0.468 | 0.496 | 0.456 | 0.544 | 1.087 | 69 | 0.6 | 0.50 |
| 0.496 | 0.525 | 0.454 | 0.546 | 1.093 | 70 | 0.6 | 0.50 |
| 0.525 | 0.554 | 0.451 | 0.549 | 1.098 | 71 | 0.6 | 0.50 |
| 0.554 | 0.583 | 0.448 | 0.552 | 1.103 | 72 | 0.6 | 0.50 |
| 0.583 | 0.613 | 0.446 | 0.554 | 1.108 | 73 | 0.6 | 0.50 |
| 0.613 | 0.643 | 0.443 | 0.557 | 1.114 | 74 | 0.6 | 0.50 |
| 0.643 | 0.675 | 0.440 | 0.560 | 1.119 | 75 | 0.6 | 0.50 |
| 0.675 | 0.706 | 0.437 | 0.563 | 1.125 | 76 | 0.6 | 0.50 |
| 0.706 | 0.739 | 0.435 | 0.565 | 1.131 | 77 | 0.6 | 0.50 |
| 0.739 | 0.772 | 0.432 | 0.568 | 1.137 | 78 | 0.6 | 0.50 |
| 0.772 | 0.807 | 0.429 | 0.571 | 1.143 | 79 | 0.6 | 0.50 |
| 0.807 | 0.842 | 0.426 | 0.574 | 1.149 | 80 | 0.6 | 0.50 |
| 0.842 | 0.878 | 0.422 | 0.578 | 1.155 | 81 | 0.6 | 0.50 |
| 0.878 | 0.916 | 0.419 | 0.581 | 1.162 | 82 | 0.6 | 0.50 |
| 0.916 | 0.954 | 0.416 | 0.584 | 1.169 | 83 | 0.6 | 0.50 |
| 0.954 | 0.995 | 0.412 | 0.588 | 1.175 | 84 | 0.6 | 0.50 |
| 0.995 | 1.037 | 0.409 | 0.591 | 1.183 | 85 | 0.6 | 0.50 |
| 1.037 | 1.080 | 0.405 | 0.595 | 1.190 | 86 | 0.6 | 0.50 |
| 1.080 | 1.127 | 0.401 | 0.599 | 1.198 | 87 | 0.6 | 0.50 |
| 1.127 | 1.175 | 0.397 | 0.603 | 1.206 | 88 | 0.6 | 0.50 |
| 1.175 | 1.227 | 0.392 | 0.608 | 1.215 | 89 | 0.6 | 0.50 |
| 1.227 | 1.282 | 0.388 | 0.612 | 1.224 | 90 | 0.6 | 0.50 |
| 1.282 | 1.341 | 0.383 | 0.617 | 1.234 | 91 | 0.6 | 0.50 |
| 1.341 | 1.405 | 0.378 | 0.622 | 1.245 | 92 | 0.6 | 0.50 |
| 1.405 | 1.476 | 0.372 | 0.628 | 1.256 | 93 | 0.6 | 0.50 |
| 1.476 | 1.555 | 0.366 | 0.634 | 1.269 | 94 | 0.6 | 0.50 |
| 1.555 | 1.645 | 0.358 | 0.642 | 1.283 | 95 | 0.6 | 0.50 |
| 1.645 | 1.751 | 0.350 | 0.650 | 1.299 | 96 | 0.6 | 0.50 |
| 1.751 | 1.881 | 0.341 | 0.659 | 1.318 | 97 | 0.6 | 0.50 |
| 1.881 | 2.054 | 0.329 | 0.671 | 1.343 | 98 | 0.6 | 0.50 |
| 2.054 | 2.326 | 0.312 | 0.688 | 1.377 | 99 | 0.6 | 0.50 |
| 2.326 | Inf | 0.276 | 0.724 | 1.449 | 100 | 0.6 | 0.50 |
| -Inf | -2.319 | 0.889 | 0.111 | 0.223 | 1 | 0.7 | 0.50 |
| -2.319 | -2.050 | 0.848 | 0.152 | 0.305 | 2 | 0.7 | 0.50 |
| -2.050 | -1.879 | 0.825 | 0.175 | 0.350 | 3 | 0.7 | 0.50 |
| -1.879 | -1.750 | 0.807 | 0.193 | 0.385 | 4 | 0.7 | 0.50 |
| -1.750 | -1.645 | 0.793 | 0.207 | 0.415 | 5 | 0.7 | 0.50 |
| -1.645 | -1.556 | 0.780 | 0.220 | 0.440 | 6 | 0.7 | 0.50 |
| -1.556 | -1.477 | 0.768 | 0.232 | 0.463 | 7 | 0.7 | 0.50 |
| -1.477 | -1.407 | 0.758 | 0.242 | 0.485 | 8 | 0.7 | 0.50 |
| -1.407 | -1.343 | 0.748 | 0.252 | 0.504 | 9 | 0.7 | 0.50 |
| -1.343 | -1.284 | 0.739 | 0.261 | 0.523 | 10 | 0.7 | 0.50 |
| -1.284 | -1.229 | 0.730 | 0.270 | 0.540 | 11 | 0.7 | 0.50 |
| -1.229 | -1.177 | 0.721 | 0.279 | 0.557 | 12 | 0.7 | 0.50 |
| -1.177 | -1.129 | 0.713 | 0.287 | 0.573 | 13 | 0.7 | 0.50 |
| -1.129 | -1.083 | 0.706 | 0.294 | 0.589 | 14 | 0.7 | 0.50 |
| -1.083 | -1.039 | 0.698 | 0.302 | 0.603 | 15 | 0.7 | 0.50 |
| -1.039 | -0.997 | 0.691 | 0.309 | 0.618 | 16 | 0.7 | 0.50 |
| -0.997 | -0.957 | 0.684 | 0.316 | 0.632 | 17 | 0.7 | 0.50 |
| -0.957 | -0.918 | 0.677 | 0.323 | 0.645 | 18 | 0.7 | 0.50 |
| -0.918 | -0.881 | 0.671 | 0.329 | 0.659 | 19 | 0.7 | 0.50 |
| -0.881 | -0.844 | 0.664 | 0.336 | 0.672 | 20 | 0.7 | 0.50 |
| -0.844 | -0.809 | 0.658 | 0.342 | 0.684 | 21 | 0.7 | 0.50 |
| -0.809 | -0.775 | 0.652 | 0.348 | 0.697 | 22 | 0.7 | 0.50 |
| -0.775 | -0.741 | 0.646 | 0.354 | 0.709 | 23 | 0.7 | 0.50 |
| -0.741 | -0.709 | 0.640 | 0.360 | 0.721 | 24 | 0.7 | 0.50 |
| -0.709 | -0.677 | 0.634 | 0.366 | 0.733 | 25 | 0.7 | 0.50 |
| -0.677 | -0.646 | 0.628 | 0.372 | 0.744 | 26 | 0.7 | 0.50 |
| -0.646 | -0.615 | 0.622 | 0.378 | 0.756 | 27 | 0.7 | 0.50 |
| -0.615 | -0.585 | 0.616 | 0.384 | 0.767 | 28 | 0.7 | 0.50 |
| -0.585 | -0.555 | 0.611 | 0.389 | 0.778 | 29 | 0.7 | 0.50 |
| -0.555 | -0.526 | 0.605 | 0.395 | 0.789 | 30 | 0.7 | 0.50 |
| -0.526 | -0.498 | 0.600 | 0.400 | 0.800 | 31 | 0.7 | 0.50 |
| -0.498 | -0.470 | 0.594 | 0.406 | 0.811 | 32 | 0.7 | 0.50 |
| -0.470 | -0.442 | 0.589 | 0.411 | 0.822 | 33 | 0.7 | 0.50 |
| -0.442 | -0.414 | 0.584 | 0.416 | 0.832 | 34 | 0.7 | 0.50 |
| -0.414 | -0.387 | 0.579 | 0.421 | 0.843 | 35 | 0.7 | 0.50 |
| -0.387 | -0.360 | 0.573 | 0.427 | 0.853 | 36 | 0.7 | 0.50 |
| -0.360 | -0.333 | 0.568 | 0.432 | 0.864 | 37 | 0.7 | 0.50 |
| -0.333 | -0.307 | 0.563 | 0.437 | 0.874 | 38 | 0.7 | 0.50 |
| -0.307 | -0.280 | 0.558 | 0.442 | 0.884 | 39 | 0.7 | 0.50 |
| -0.280 | -0.254 | 0.553 | 0.447 | 0.895 | 40 | 0.7 | 0.50 |
| -0.254 | -0.229 | 0.548 | 0.452 | 0.905 | 41 | 0.7 | 0.50 |
| -0.229 | -0.203 | 0.543 | 0.457 | 0.915 | 42 | 0.7 | 0.50 |
| -0.203 | -0.177 | 0.537 | 0.463 | 0.925 | 43 | 0.7 | 0.50 |
| -0.177 | -0.152 | 0.532 | 0.468 | 0.935 | 44 | 0.7 | 0.50 |
| -0.152 | -0.126 | 0.527 | 0.473 | 0.945 | 45 | 0.7 | 0.50 |
| -0.126 | -0.101 | 0.522 | 0.478 | 0.955 | 46 | 0.7 | 0.50 |
| -0.101 | -0.076 | 0.517 | 0.483 | 0.965 | 47 | 0.7 | 0.50 |
| -0.076 | -0.050 | 0.512 | 0.488 | 0.975 | 48 | 0.7 | 0.50 |
| -0.050 | -0.025 | 0.507 | 0.493 | 0.985 | 49 | 0.7 | 0.50 |
| -0.025 | 0.000 | 0.502 | 0.498 | 0.995 | 50 | 0.7 | 0.50 |
| 0.000 | 0.025 | 0.498 | 0.502 | 1.005 | 51 | 0.7 | 0.50 |
| 0.025 | 0.050 | 0.493 | 0.507 | 1.015 | 52 | 0.7 | 0.50 |
| 0.050 | 0.076 | 0.488 | 0.512 | 1.025 | 53 | 0.7 | 0.50 |
| 0.076 | 0.101 | 0.483 | 0.517 | 1.035 | 54 | 0.7 | 0.50 |
| 0.101 | 0.126 | 0.478 | 0.522 | 1.045 | 55 | 0.7 | 0.50 |
| 0.126 | 0.152 | 0.473 | 0.527 | 1.055 | 56 | 0.7 | 0.50 |
| 0.152 | 0.177 | 0.468 | 0.532 | 1.065 | 57 | 0.7 | 0.50 |
| 0.177 | 0.203 | 0.463 | 0.537 | 1.075 | 58 | 0.7 | 0.50 |
| 0.203 | 0.229 | 0.457 | 0.543 | 1.085 | 59 | 0.7 | 0.50 |
| 0.229 | 0.254 | 0.452 | 0.548 | 1.095 | 60 | 0.7 | 0.50 |
| 0.254 | 0.280 | 0.447 | 0.553 | 1.105 | 61 | 0.7 | 0.50 |
| 0.280 | 0.307 | 0.442 | 0.558 | 1.116 | 62 | 0.7 | 0.50 |
| 0.307 | 0.333 | 0.437 | 0.563 | 1.126 | 63 | 0.7 | 0.50 |
| 0.333 | 0.360 | 0.432 | 0.568 | 1.136 | 64 | 0.7 | 0.50 |
| 0.360 | 0.387 | 0.427 | 0.573 | 1.147 | 65 | 0.7 | 0.50 |
| 0.387 | 0.414 | 0.421 | 0.579 | 1.157 | 66 | 0.7 | 0.50 |
| 0.414 | 0.442 | 0.416 | 0.584 | 1.168 | 67 | 0.7 | 0.50 |
| 0.442 | 0.470 | 0.411 | 0.589 | 1.178 | 68 | 0.7 | 0.50 |
| 0.470 | 0.498 | 0.406 | 0.594 | 1.189 | 69 | 0.7 | 0.50 |
| 0.498 | 0.526 | 0.400 | 0.600 | 1.200 | 70 | 0.7 | 0.50 |
| 0.526 | 0.555 | 0.395 | 0.605 | 1.211 | 71 | 0.7 | 0.50 |
| 0.555 | 0.585 | 0.389 | 0.611 | 1.222 | 72 | 0.7 | 0.50 |
| 0.585 | 0.615 | 0.384 | 0.616 | 1.233 | 73 | 0.7 | 0.50 |
| 0.615 | 0.646 | 0.378 | 0.622 | 1.244 | 74 | 0.7 | 0.50 |
| 0.646 | 0.677 | 0.372 | 0.628 | 1.256 | 75 | 0.7 | 0.50 |
| 0.677 | 0.709 | 0.366 | 0.634 | 1.267 | 76 | 0.7 | 0.50 |
| 0.709 | 0.741 | 0.360 | 0.640 | 1.279 | 77 | 0.7 | 0.50 |
| 0.741 | 0.775 | 0.354 | 0.646 | 1.291 | 78 | 0.7 | 0.50 |
| 0.775 | 0.809 | 0.348 | 0.652 | 1.303 | 79 | 0.7 | 0.50 |
| 0.809 | 0.844 | 0.342 | 0.658 | 1.316 | 80 | 0.7 | 0.50 |
| 0.844 | 0.881 | 0.336 | 0.664 | 1.328 | 81 | 0.7 | 0.50 |
| 0.881 | 0.918 | 0.329 | 0.671 | 1.341 | 82 | 0.7 | 0.50 |
| 0.918 | 0.957 | 0.323 | 0.677 | 1.355 | 83 | 0.7 | 0.50 |
| 0.957 | 0.997 | 0.316 | 0.684 | 1.368 | 84 | 0.7 | 0.50 |
| 0.997 | 1.039 | 0.309 | 0.691 | 1.382 | 85 | 0.7 | 0.50 |
| 1.039 | 1.083 | 0.302 | 0.698 | 1.397 | 86 | 0.7 | 0.50 |
| 1.083 | 1.129 | 0.294 | 0.706 | 1.411 | 87 | 0.7 | 0.50 |
| 1.129 | 1.177 | 0.287 | 0.713 | 1.427 | 88 | 0.7 | 0.50 |
| 1.177 | 1.229 | 0.279 | 0.721 | 1.443 | 89 | 0.7 | 0.50 |
| 1.229 | 1.284 | 0.270 | 0.730 | 1.460 | 90 | 0.7 | 0.50 |
| 1.284 | 1.343 | 0.261 | 0.739 | 1.477 | 91 | 0.7 | 0.50 |
| 1.343 | 1.407 | 0.252 | 0.748 | 1.496 | 92 | 0.7 | 0.50 |
| 1.407 | 1.477 | 0.242 | 0.758 | 1.515 | 93 | 0.7 | 0.50 |
| 1.477 | 1.556 | 0.232 | 0.768 | 1.537 | 94 | 0.7 | 0.50 |
| 1.556 | 1.645 | 0.220 | 0.780 | 1.560 | 95 | 0.7 | 0.50 |
| 1.645 | 1.750 | 0.207 | 0.793 | 1.585 | 96 | 0.7 | 0.50 |
| 1.750 | 1.879 | 0.193 | 0.807 | 1.615 | 97 | 0.7 | 0.50 |
| 1.879 | 2.050 | 0.175 | 0.825 | 1.650 | 98 | 0.7 | 0.50 |
| 2.050 | 2.319 | 0.152 | 0.848 | 1.695 | 99 | 0.7 | 0.50 |
| 2.319 | Inf | 0.111 | 0.889 | 1.777 | 100 | 0.7 | 0.50 |
| -Inf | -2.286 | 0.972 | 0.028 | 0.057 | 1 | 0.8 | 0.50 |
| -2.286 | -2.031 | 0.951 | 0.049 | 0.098 | 2 | 0.8 | 0.50 |
| -2.031 | -1.868 | 0.937 | 0.063 | 0.127 | 3 | 0.8 | 0.50 |
| -1.868 | -1.744 | 0.924 | 0.076 | 0.152 | 4 | 0.8 | 0.50 |
| -1.744 | -1.643 | 0.912 | 0.088 | 0.175 | 5 | 0.8 | 0.50 |
| -1.643 | -1.557 | 0.902 | 0.098 | 0.197 | 6 | 0.8 | 0.50 |
| -1.557 | -1.480 | 0.891 | 0.109 | 0.218 | 7 | 0.8 | 0.50 |
| -1.480 | -1.412 | 0.881 | 0.119 | 0.238 | 8 | 0.8 | 0.50 |
| -1.412 | -1.349 | 0.871 | 0.129 | 0.258 | 9 | 0.8 | 0.50 |
| -1.349 | -1.291 | 0.862 | 0.138 | 0.277 | 10 | 0.8 | 0.50 |
| -1.291 | -1.237 | 0.852 | 0.148 | 0.296 | 11 | 0.8 | 0.50 |
| -1.237 | -1.187 | 0.843 | 0.157 | 0.315 | 12 | 0.8 | 0.50 |
| -1.187 | -1.139 | 0.833 | 0.167 | 0.333 | 13 | 0.8 | 0.50 |
| -1.139 | -1.093 | 0.824 | 0.176 | 0.351 | 14 | 0.8 | 0.50 |
| -1.093 | -1.050 | 0.815 | 0.185 | 0.370 | 15 | 0.8 | 0.50 |
| -1.050 | -1.008 | 0.806 | 0.194 | 0.388 | 16 | 0.8 | 0.50 |
| -1.008 | -0.968 | 0.797 | 0.203 | 0.406 | 17 | 0.8 | 0.50 |
| -0.968 | -0.930 | 0.788 | 0.212 | 0.424 | 18 | 0.8 | 0.50 |
| -0.930 | -0.892 | 0.779 | 0.221 | 0.441 | 19 | 0.8 | 0.50 |
| -0.892 | -0.856 | 0.770 | 0.230 | 0.459 | 20 | 0.8 | 0.50 |
| -0.856 | -0.821 | 0.762 | 0.238 | 0.477 | 21 | 0.8 | 0.50 |
| -0.821 | -0.786 | 0.753 | 0.247 | 0.495 | 22 | 0.8 | 0.50 |
| -0.786 | -0.753 | 0.744 | 0.256 | 0.512 | 23 | 0.8 | 0.50 |
| -0.753 | -0.720 | 0.735 | 0.265 | 0.530 | 24 | 0.8 | 0.50 |
| -0.720 | -0.688 | 0.726 | 0.274 | 0.548 | 25 | 0.8 | 0.50 |
| -0.688 | -0.657 | 0.717 | 0.283 | 0.565 | 26 | 0.8 | 0.50 |
| -0.657 | -0.626 | 0.708 | 0.292 | 0.583 | 27 | 0.8 | 0.50 |
| -0.626 | -0.595 | 0.700 | 0.300 | 0.601 | 28 | 0.8 | 0.50 |
| -0.595 | -0.565 | 0.691 | 0.309 | 0.618 | 29 | 0.8 | 0.50 |
| -0.565 | -0.536 | 0.682 | 0.318 | 0.636 | 30 | 0.8 | 0.50 |
| -0.536 | -0.507 | 0.673 | 0.327 | 0.654 | 31 | 0.8 | 0.50 |
| -0.507 | -0.478 | 0.664 | 0.336 | 0.671 | 32 | 0.8 | 0.50 |
| -0.478 | -0.450 | 0.655 | 0.345 | 0.689 | 33 | 0.8 | 0.50 |
| -0.450 | -0.422 | 0.647 | 0.353 | 0.707 | 34 | 0.8 | 0.50 |
| -0.422 | -0.395 | 0.638 | 0.362 | 0.725 | 35 | 0.8 | 0.50 |
| -0.395 | -0.367 | 0.629 | 0.371 | 0.742 | 36 | 0.8 | 0.50 |
| -0.367 | -0.340 | 0.620 | 0.380 | 0.760 | 37 | 0.8 | 0.50 |
| -0.340 | -0.313 | 0.611 | 0.389 | 0.778 | 38 | 0.8 | 0.50 |
| -0.313 | -0.286 | 0.602 | 0.398 | 0.795 | 39 | 0.8 | 0.50 |
| -0.286 | -0.260 | 0.593 | 0.407 | 0.813 | 40 | 0.8 | 0.50 |
| -0.260 | -0.233 | 0.585 | 0.415 | 0.831 | 41 | 0.8 | 0.50 |
| -0.233 | -0.207 | 0.576 | 0.424 | 0.849 | 42 | 0.8 | 0.50 |
| -0.207 | -0.181 | 0.567 | 0.433 | 0.867 | 43 | 0.8 | 0.50 |
| -0.181 | -0.155 | 0.558 | 0.442 | 0.884 | 44 | 0.8 | 0.50 |
| -0.155 | -0.129 | 0.549 | 0.451 | 0.902 | 45 | 0.8 | 0.50 |
| -0.129 | -0.103 | 0.540 | 0.460 | 0.920 | 46 | 0.8 | 0.50 |
| -0.103 | -0.077 | 0.531 | 0.469 | 0.938 | 47 | 0.8 | 0.50 |
| -0.077 | -0.051 | 0.522 | 0.478 | 0.955 | 48 | 0.8 | 0.50 |
| -0.051 | -0.026 | 0.513 | 0.487 | 0.973 | 49 | 0.8 | 0.50 |
| -0.026 | 0.000 | 0.504 | 0.496 | 0.991 | 50 | 0.8 | 0.50 |
| 0.000 | 0.026 | 0.496 | 0.504 | 1.009 | 51 | 0.8 | 0.50 |
| 0.026 | 0.051 | 0.487 | 0.513 | 1.027 | 52 | 0.8 | 0.50 |
| 0.051 | 0.077 | 0.478 | 0.522 | 1.045 | 53 | 0.8 | 0.50 |
| 0.077 | 0.103 | 0.469 | 0.531 | 1.062 | 54 | 0.8 | 0.50 |
| 0.103 | 0.129 | 0.460 | 0.540 | 1.080 | 55 | 0.8 | 0.50 |
| 0.129 | 0.155 | 0.451 | 0.549 | 1.098 | 56 | 0.8 | 0.50 |
| 0.155 | 0.181 | 0.442 | 0.558 | 1.116 | 57 | 0.8 | 0.50 |
| 0.181 | 0.207 | 0.433 | 0.567 | 1.133 | 58 | 0.8 | 0.50 |
| 0.207 | 0.233 | 0.424 | 0.576 | 1.151 | 59 | 0.8 | 0.50 |
| 0.233 | 0.260 | 0.415 | 0.585 | 1.169 | 60 | 0.8 | 0.50 |
| 0.260 | 0.286 | 0.407 | 0.593 | 1.187 | 61 | 0.8 | 0.50 |
| 0.286 | 0.313 | 0.398 | 0.602 | 1.205 | 62 | 0.8 | 0.50 |
| 0.313 | 0.340 | 0.389 | 0.611 | 1.222 | 63 | 0.8 | 0.50 |
| 0.340 | 0.367 | 0.380 | 0.620 | 1.240 | 64 | 0.8 | 0.50 |
| 0.367 | 0.395 | 0.371 | 0.629 | 1.258 | 65 | 0.8 | 0.50 |
| 0.395 | 0.422 | 0.362 | 0.638 | 1.275 | 66 | 0.8 | 0.50 |
| 0.422 | 0.450 | 0.353 | 0.647 | 1.293 | 67 | 0.8 | 0.50 |
| 0.450 | 0.478 | 0.345 | 0.655 | 1.311 | 68 | 0.8 | 0.50 |
| 0.478 | 0.507 | 0.336 | 0.664 | 1.329 | 69 | 0.8 | 0.50 |
| 0.507 | 0.536 | 0.327 | 0.673 | 1.346 | 70 | 0.8 | 0.50 |
| 0.536 | 0.565 | 0.318 | 0.682 | 1.364 | 71 | 0.8 | 0.50 |
| 0.565 | 0.595 | 0.309 | 0.691 | 1.382 | 72 | 0.8 | 0.50 |
| 0.595 | 0.626 | 0.300 | 0.700 | 1.399 | 73 | 0.8 | 0.50 |
| 0.626 | 0.657 | 0.292 | 0.708 | 1.417 | 74 | 0.8 | 0.50 |
| 0.657 | 0.688 | 0.283 | 0.717 | 1.435 | 75 | 0.8 | 0.50 |
| 0.688 | 0.720 | 0.274 | 0.726 | 1.452 | 76 | 0.8 | 0.50 |
| 0.720 | 0.753 | 0.265 | 0.735 | 1.470 | 77 | 0.8 | 0.50 |
| 0.753 | 0.786 | 0.256 | 0.744 | 1.488 | 78 | 0.8 | 0.50 |
| 0.786 | 0.821 | 0.247 | 0.753 | 1.505 | 79 | 0.8 | 0.50 |
| 0.821 | 0.856 | 0.238 | 0.762 | 1.523 | 80 | 0.8 | 0.50 |
| 0.856 | 0.892 | 0.230 | 0.770 | 1.541 | 81 | 0.8 | 0.50 |
| 0.892 | 0.930 | 0.221 | 0.779 | 1.559 | 82 | 0.8 | 0.50 |
| 0.930 | 0.968 | 0.212 | 0.788 | 1.576 | 83 | 0.8 | 0.50 |
| 0.968 | 1.008 | 0.203 | 0.797 | 1.594 | 84 | 0.8 | 0.50 |
| 1.008 | 1.050 | 0.194 | 0.806 | 1.612 | 85 | 0.8 | 0.50 |
| 1.050 | 1.093 | 0.185 | 0.815 | 1.630 | 86 | 0.8 | 0.50 |
| 1.093 | 1.139 | 0.176 | 0.824 | 1.649 | 87 | 0.8 | 0.50 |
| 1.139 | 1.187 | 0.167 | 0.833 | 1.667 | 88 | 0.8 | 0.50 |
| 1.187 | 1.237 | 0.157 | 0.843 | 1.685 | 89 | 0.8 | 0.50 |
| 1.237 | 1.291 | 0.148 | 0.852 | 1.704 | 90 | 0.8 | 0.50 |
| 1.291 | 1.349 | 0.138 | 0.862 | 1.723 | 91 | 0.8 | 0.50 |
| 1.349 | 1.412 | 0.129 | 0.871 | 1.742 | 92 | 0.8 | 0.50 |
| 1.412 | 1.480 | 0.119 | 0.881 | 1.762 | 93 | 0.8 | 0.50 |
| 1.480 | 1.557 | 0.109 | 0.891 | 1.782 | 94 | 0.8 | 0.50 |
| 1.557 | 1.643 | 0.098 | 0.902 | 1.803 | 95 | 0.8 | 0.50 |
| 1.643 | 1.744 | 0.088 | 0.912 | 1.825 | 96 | 0.8 | 0.50 |
| 1.744 | 1.868 | 0.076 | 0.924 | 1.848 | 97 | 0.8 | 0.50 |
| 1.868 | 2.031 | 0.063 | 0.937 | 1.873 | 98 | 0.8 | 0.50 |
| 2.031 | 2.286 | 0.049 | 0.951 | 1.902 | 99 | 0.8 | 0.50 |
| 2.286 | Inf | 0.028 | 0.972 | 1.943 | 100 | 0.8 | 0.50 |
| -Inf | -2.194 | 0.997 | 0.003 | 0.006 | 1 | 0.9 | 0.50 |
| -2.194 | -1.970 | 0.994 | 0.006 | 0.013 | 2 | 0.9 | 0.50 |
| -1.970 | -1.826 | 0.990 | 0.010 | 0.019 | 3 | 0.9 | 0.50 |
| -1.826 | -1.716 | 0.987 | 0.013 | 0.026 | 4 | 0.9 | 0.50 |
| -1.716 | -1.625 | 0.983 | 0.017 | 0.033 | 5 | 0.9 | 0.50 |
| -1.625 | -1.547 | 0.980 | 0.020 | 0.041 | 6 | 0.9 | 0.50 |
| -1.547 | -1.478 | 0.976 | 0.024 | 0.048 | 7 | 0.9 | 0.50 |
| -1.478 | -1.416 | 0.972 | 0.028 | 0.057 | 8 | 0.9 | 0.50 |
| -1.416 | -1.359 | 0.967 | 0.033 | 0.065 | 9 | 0.9 | 0.50 |
| -1.359 | -1.305 | 0.963 | 0.037 | 0.074 | 10 | 0.9 | 0.50 |
| -1.305 | -1.255 | 0.958 | 0.042 | 0.084 | 11 | 0.9 | 0.50 |
| -1.255 | -1.208 | 0.953 | 0.047 | 0.094 | 12 | 0.9 | 0.50 |
| -1.208 | -1.163 | 0.948 | 0.052 | 0.104 | 13 | 0.9 | 0.50 |
| -1.163 | -1.121 | 0.942 | 0.058 | 0.115 | 14 | 0.9 | 0.50 |
| -1.121 | -1.080 | 0.936 | 0.064 | 0.127 | 15 | 0.9 | 0.50 |
| -1.080 | -1.040 | 0.930 | 0.070 | 0.139 | 16 | 0.9 | 0.50 |
| -1.040 | -1.002 | 0.924 | 0.076 | 0.152 | 17 | 0.9 | 0.50 |
| -1.002 | -0.964 | 0.917 | 0.083 | 0.166 | 18 | 0.9 | 0.50 |
| -0.964 | -0.928 | 0.910 | 0.090 | 0.180 | 19 | 0.9 | 0.50 |
| -0.928 | -0.893 | 0.903 | 0.097 | 0.195 | 20 | 0.9 | 0.50 |
| -0.893 | -0.858 | 0.895 | 0.105 | 0.210 | 21 | 0.9 | 0.50 |
| -0.858 | -0.824 | 0.887 | 0.113 | 0.227 | 22 | 0.9 | 0.50 |
| -0.824 | -0.791 | 0.878 | 0.122 | 0.244 | 23 | 0.9 | 0.50 |
| -0.791 | -0.759 | 0.869 | 0.131 | 0.261 | 24 | 0.9 | 0.50 |
| -0.759 | -0.727 | 0.860 | 0.140 | 0.280 | 25 | 0.9 | 0.50 |
| -0.727 | -0.695 | 0.850 | 0.150 | 0.299 | 26 | 0.9 | 0.50 |
| -0.695 | -0.664 | 0.840 | 0.160 | 0.319 | 27 | 0.9 | 0.50 |
| -0.664 | -0.633 | 0.830 | 0.170 | 0.340 | 28 | 0.9 | 0.50 |
| -0.633 | -0.602 | 0.819 | 0.181 | 0.362 | 29 | 0.9 | 0.50 |
| -0.602 | -0.572 | 0.808 | 0.192 | 0.384 | 30 | 0.9 | 0.50 |
| -0.572 | -0.542 | 0.796 | 0.204 | 0.408 | 31 | 0.9 | 0.50 |
| -0.542 | -0.512 | 0.784 | 0.216 | 0.432 | 32 | 0.9 | 0.50 |
| -0.512 | -0.483 | 0.772 | 0.228 | 0.457 | 33 | 0.9 | 0.50 |
| -0.483 | -0.454 | 0.759 | 0.241 | 0.483 | 34 | 0.9 | 0.50 |
| -0.454 | -0.425 | 0.745 | 0.255 | 0.509 | 35 | 0.9 | 0.50 |
| -0.425 | -0.396 | 0.732 | 0.268 | 0.537 | 36 | 0.9 | 0.50 |
| -0.396 | -0.367 | 0.718 | 0.282 | 0.565 | 37 | 0.9 | 0.50 |
| -0.367 | -0.338 | 0.703 | 0.297 | 0.594 | 38 | 0.9 | 0.50 |
| -0.338 | -0.310 | 0.688 | 0.312 | 0.623 | 39 | 0.9 | 0.50 |
| -0.310 | -0.281 | 0.673 | 0.327 | 0.654 | 40 | 0.9 | 0.50 |
| -0.281 | -0.253 | 0.658 | 0.342 | 0.684 | 41 | 0.9 | 0.50 |
| -0.253 | -0.225 | 0.642 | 0.358 | 0.716 | 42 | 0.9 | 0.50 |
| -0.225 | -0.196 | 0.626 | 0.374 | 0.748 | 43 | 0.9 | 0.50 |
| -0.196 | -0.168 | 0.610 | 0.390 | 0.781 | 44 | 0.9 | 0.50 |
| -0.168 | -0.140 | 0.593 | 0.407 | 0.814 | 45 | 0.9 | 0.50 |
| -0.140 | -0.112 | 0.577 | 0.423 | 0.847 | 46 | 0.9 | 0.50 |
| -0.112 | -0.084 | 0.560 | 0.440 | 0.881 | 47 | 0.9 | 0.50 |
| -0.084 | -0.056 | 0.543 | 0.457 | 0.915 | 48 | 0.9 | 0.50 |
| -0.056 | -0.028 | 0.526 | 0.474 | 0.949 | 49 | 0.9 | 0.50 |
| -0.028 | 0.000 | 0.509 | 0.491 | 0.983 | 50 | 0.9 | 0.50 |
| 0.000 | 0.028 | 0.491 | 0.509 | 1.017 | 51 | 0.9 | 0.50 |
| 0.028 | 0.056 | 0.474 | 0.526 | 1.051 | 52 | 0.9 | 0.50 |
| 0.056 | 0.084 | 0.457 | 0.543 | 1.085 | 53 | 0.9 | 0.50 |
| 0.084 | 0.112 | 0.440 | 0.560 | 1.119 | 54 | 0.9 | 0.50 |
| 0.112 | 0.140 | 0.423 | 0.577 | 1.153 | 55 | 0.9 | 0.50 |
| 0.140 | 0.168 | 0.407 | 0.593 | 1.186 | 56 | 0.9 | 0.50 |
| 0.168 | 0.196 | 0.390 | 0.610 | 1.219 | 57 | 0.9 | 0.50 |
| 0.196 | 0.225 | 0.374 | 0.626 | 1.252 | 58 | 0.9 | 0.50 |
| 0.225 | 0.253 | 0.358 | 0.642 | 1.284 | 59 | 0.9 | 0.50 |
| 0.253 | 0.281 | 0.342 | 0.658 | 1.316 | 60 | 0.9 | 0.50 |
| 0.281 | 0.310 | 0.327 | 0.673 | 1.346 | 61 | 0.9 | 0.50 |
| 0.310 | 0.338 | 0.312 | 0.688 | 1.377 | 62 | 0.9 | 0.50 |
| 0.338 | 0.367 | 0.297 | 0.703 | 1.406 | 63 | 0.9 | 0.50 |
| 0.367 | 0.396 | 0.282 | 0.718 | 1.435 | 64 | 0.9 | 0.50 |
| 0.396 | 0.425 | 0.268 | 0.732 | 1.463 | 65 | 0.9 | 0.50 |
| 0.425 | 0.454 | 0.255 | 0.745 | 1.491 | 66 | 0.9 | 0.50 |
| 0.454 | 0.483 | 0.241 | 0.759 | 1.517 | 67 | 0.9 | 0.50 |
| 0.483 | 0.512 | 0.228 | 0.772 | 1.543 | 68 | 0.9 | 0.50 |
| 0.512 | 0.542 | 0.216 | 0.784 | 1.568 | 69 | 0.9 | 0.50 |
| 0.542 | 0.572 | 0.204 | 0.796 | 1.592 | 70 | 0.9 | 0.50 |
| 0.572 | 0.602 | 0.192 | 0.808 | 1.616 | 71 | 0.9 | 0.50 |
| 0.602 | 0.633 | 0.181 | 0.819 | 1.638 | 72 | 0.9 | 0.50 |
| 0.633 | 0.664 | 0.170 | 0.830 | 1.660 | 73 | 0.9 | 0.50 |
| 0.664 | 0.695 | 0.160 | 0.840 | 1.681 | 74 | 0.9 | 0.50 |
| 0.695 | 0.727 | 0.150 | 0.850 | 1.701 | 75 | 0.9 | 0.50 |
| 0.727 | 0.759 | 0.140 | 0.860 | 1.720 | 76 | 0.9 | 0.50 |
| 0.759 | 0.791 | 0.131 | 0.869 | 1.739 | 77 | 0.9 | 0.50 |
| 0.791 | 0.824 | 0.122 | 0.878 | 1.756 | 78 | 0.9 | 0.50 |
| 0.824 | 0.858 | 0.113 | 0.887 | 1.773 | 79 | 0.9 | 0.50 |
| 0.858 | 0.893 | 0.105 | 0.895 | 1.790 | 80 | 0.9 | 0.50 |
| 0.893 | 0.928 | 0.097 | 0.903 | 1.805 | 81 | 0.9 | 0.50 |
| 0.928 | 0.964 | 0.090 | 0.910 | 1.820 | 82 | 0.9 | 0.50 |
| 0.964 | 1.002 | 0.083 | 0.917 | 1.834 | 83 | 0.9 | 0.50 |
| 1.002 | 1.040 | 0.076 | 0.924 | 1.848 | 84 | 0.9 | 0.50 |
| 1.040 | 1.080 | 0.070 | 0.930 | 1.861 | 85 | 0.9 | 0.50 |
| 1.080 | 1.121 | 0.064 | 0.936 | 1.873 | 86 | 0.9 | 0.50 |
| 1.121 | 1.163 | 0.058 | 0.942 | 1.885 | 87 | 0.9 | 0.50 |
| 1.163 | 1.208 | 0.052 | 0.948 | 1.896 | 88 | 0.9 | 0.50 |
| 1.208 | 1.255 | 0.047 | 0.953 | 1.906 | 89 | 0.9 | 0.50 |
| 1.255 | 1.305 | 0.042 | 0.958 | 1.916 | 90 | 0.9 | 0.50 |
| 1.305 | 1.359 | 0.037 | 0.963 | 1.926 | 91 | 0.9 | 0.50 |
| 1.359 | 1.416 | 0.033 | 0.967 | 1.935 | 92 | 0.9 | 0.50 |
| 1.416 | 1.478 | 0.028 | 0.972 | 1.943 | 93 | 0.9 | 0.50 |
| 1.478 | 1.547 | 0.024 | 0.976 | 1.952 | 94 | 0.9 | 0.50 |
| 1.547 | 1.625 | 0.020 | 0.980 | 1.959 | 95 | 0.9 | 0.50 |
| 1.625 | 1.716 | 0.017 | 0.983 | 1.967 | 96 | 0.9 | 0.50 |
| 1.716 | 1.826 | 0.013 | 0.987 | 1.974 | 97 | 0.9 | 0.50 |
| 1.826 | 1.970 | 0.010 | 0.990 | 1.981 | 98 | 0.9 | 0.50 |
| 1.970 | 2.194 | 0.006 | 0.994 | 1.987 | 99 | 0.9 | 0.50 |
| 2.194 | Inf | 0.003 | 0.997 | 1.994 | 100 | 0.9 | 0.50 |
1.4 Interactive tool for converting PRS into absolute risk estimates
1.4.1 Binary outcomes
Show tool
1.4.2 Quantitative outcomes
Show tool
Both these conversions require estimates of AUC/R2 by the polygenic score. However, this information is often not known for a given polygenic score so approaches to estimate the variance explained by a polygenic scores using summary statistics is required.
2 Estimating AUC/R2 by polygenic score
Here we will use an approach using LD-score regression to estimate the SNP-based heritability of the GWAS phenotype, and subsequent use of AVENGEME to estimate the variance explained by the polygenic score given the heritability of the phenotype and the sample size of the GWAS.
We validate this method by comparison to observed variance explained measurements in UK Biobank. We use external GWAS summary statistics to generate polygenic scores.
2.1 Validation using External GWAS
Estimate the AUC/R2 of polygenic scores from external GWAS, and compare to observed estimates.
2.1.1 LDSC/AVENGEME
2.1.1.1 LDSC
Show results
# Create version of HapMap3 SNP list that doesn't contain the MHC region
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
hm3<-fread(paste0(ldsc_ref,'/w_hm3.snplist'))
bim<-fread('/scratch/groups/biomarkers-brc-mh/Reference_data/1KG_Phase3/PLINK/all_phase3.chr6.bim')
bim_mhc<-bim[bim$V1 == 6 & bim$V4 > 28e6 & bim$V4 < 34e6,]
hm3_nomhc<-hm3[!(hm3$SNP %in% bim_mhc$V2),]
write.table(hm3_nomhc, paste0(ldsc_ref,'/w_hm3_nomhc.snplist'), col.names = T, row.names = F, quote = F)
########
# Run LDSC for binary outcomes
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
pop_prev=c(0.15,0.05,0.03,0.013,0.00164,0.005,0.125,0.125)
samp_prev=c(0.318,0.168,0.33,0.592,0.36,0.246,0.537,0.564)
# Munge the sumstats
for(i in 1:length(pheno)){
system(paste0(munge_sumstats,' --sumstats ',gwas_rep_qcd,'/',gwas[i],'.cleaned.gz --merge-alleles ',ldsc_ref,'/w_hm3_nomhc.snplist --out ', gwas_rep_qcd,'/',gwas[i],'.munged'))
}
# Estimate heritability
for(i in 1:length(pheno)){
system(paste0(ldsc,' --h2 ',gwas_rep_qcd,'/',gwas[i],'.munged.sumstats.gz --ref-ld-chr ',ldsc_ref,'/ --w-ld-chr ',ldsc_ref,'/ --out /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/',pheno[i],'_h2 --samp-prev ',samp_prev[i],' --pop-prev ',pop_prev[i]))
}
h2_all<-NULL
for(i in 1:length(pheno)){
ldsc_log<-read.table(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/',pheno[i],'_h2.log'), header=F, sep='&')
ldsc_h2<-ldsc_log[grepl('Total Liability scale h2', ldsc_log$V1),]
ldsc_h2<-gsub('Total Liability scale h2: ','', ldsc_h2)
ldsc_h2_est<-as.numeric(gsub(' .*','', ldsc_h2))
ldsc_h2_se<-as.numeric(gsub("\\)",'',gsub(".*\\(",'', ldsc_h2)))
ldsc_int<-ldsc_log[grepl('Intercept: ', ldsc_log$V1),]
ldsc_int<-gsub('Intercept: ','', ldsc_int)
ldsc_int_est<-as.numeric(gsub(' .*','', ldsc_int))
ldsc_int_se<-as.numeric(gsub("\\)",'',gsub(".*\\(",'', ldsc_int)))
munged<-fread(cmd=paste0('zcat ',gwas_rep_qcd,'/',gwas[i],'.munged.sumstats.gz'))
N<-median(munged$N, na.rm = T)
h2_all<-rbind(h2_all, data.frame(GWAS=gwas[i],
Phenotype=pheno[i],
h2=ldsc_h2_est,
h2_se=ldsc_h2_se,
int=ldsc_int_est,
int_se=ldsc_int_se,
pop_prev=pop_prev[i],
samp_prev=samp_prev[i],
N=N))
}
write.csv(h2_all, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/ldsc_h2.csv', row.names=F, quote=F)
#####
# Continuous outcomes
#####
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
gwas<-c('COLL01','HEIG03','BODY04')
pheno=c('Intelligence','BMI','Height')
# Munge the sumstats
for(i in 1:length(pheno)){
system(paste0(munge_sumstats,' --sumstats ',gwas_rep_qcd,'/',gwas[i],'.cleaned.gz --merge-alleles ',ldsc_ref,'/w_hm3_nomhc.snplist --out ', gwas_rep_qcd,'/',gwas[i],'.munged'))
}
# Estimate heritability
for(i in 1:length(pheno)){
system(paste0(ldsc,' --h2 ',gwas_rep_qcd,'/',gwas[i],'.munged.sumstats.gz --ref-ld-chr ',ldsc_ref,'/ --w-ld-chr ',ldsc_ref,'/ --out /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/',pheno[i],'_h2_obs'))
}
h2_all<-NULL
for(i in 1:length(pheno)){
ldsc_log<-read.table(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/',pheno[i],'_h2_obs.log'), header=F, sep='&')
ldsc_h2<-ldsc_log[grepl('Total Observed scale h2', ldsc_log$V1),]
ldsc_h2<-gsub('Total Observed scale h2: ','', ldsc_h2)
ldsc_h2_est<-as.numeric(gsub(' .*','', ldsc_h2))
ldsc_h2_se<-as.numeric(gsub("\\)",'',gsub(".*\\(",'', ldsc_h2)))
ldsc_int<-ldsc_log[grepl('Intercept: ', ldsc_log$V1),]
ldsc_int<-gsub('Intercept: ','', ldsc_int)
ldsc_int_est<-as.numeric(gsub(' .*','', ldsc_int))
ldsc_int_se<-as.numeric(gsub("\\)",'',gsub(".*\\(",'', ldsc_int)))
munged<-fread(cmd=paste0('zcat ',gwas_rep_qcd,'/',gwas[i],'.munged.sumstats.gz'))
N<-median(munged$N, na.rm = T)
h2_all<-rbind(h2_all, data.frame(GWAS=gwas[i],
Phenotype=pheno[i],
h2=ldsc_h2_est,
h2_se=ldsc_h2_se,
int=ldsc_int_est,
int_se=ldsc_int_se,
N=N))
}
write.csv(h2_all, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/ldsc_h2_obs.csv', row.names=F, quote=F)2.1.1.2 AVENGEME
Show code
library(avengeme)
library(ggplot2)
library(cowplot)
library(data.table)
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
#####
# Estimate using the liability h2
#####
h2<-fread('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/ldsc_h2.csv')
AUC_pred<-NULL
AVENGEME_plots<-list()
for(i in 1:length(pheno)){
# Read in observed AUC estimates
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',pheno[i],'/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
# Estimate heritability using AVENGEME
pheno_nsnp<-NULL
tmp<-fread(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/pt_clump/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.NSNP_per_pT'))
pheno_nsnp<-tmp$NSNP[length(tmp$NSNP)]
pheno_res_pTclump<-assoc[grepl('PredFile1',assoc$Predictor),]
pT<-as.numeric(gsub('e.','e-',gsub('.*_','',pheno_res_pTclump$Predictor)))
pheno_res_pTclump$pT<-pT
tmp2<-estimatePolygenicModel(p=pheno_res_pTclump$Estimate/pheno_res_pTclump$SE,
nsnp=pheno_nsnp,
n=c(h2$N[h2$Phenotype == pheno[i]], pheno_res_pTclump$N[1]),
pupper = c(0,pT),
binary = c(T, T),
prevalence = h2$pop_prev[h2$Phenotype == pheno[i]],
sampling = c(h2$samp_prev[h2$Phenotype == pheno[i]], (pheno_res_pTclump$Ncas/pheno_res_pTclump$N)[1]),
fixvg2pi02 = T,
alpha = 0.05)
# Run AVENGEME predicted AUC using LDSC heritability
avengeme_res_ldsc<-NULL
for(pT_i in pT){
for(pi0 in seq(0.92,0.98,0.02)){
tmp<-polygenescore(nsnp=pheno_nsnp, n=h2$N[h2$Phenotype == pheno[i]], vg1 = h2$h2[h2$Phenotype == pheno[i]], pupper = c(0, pT_i), pi0 = pi0, nested = TRUE, weighted = TRUE, binary = T, prevalence = h2$pop_prev[h2$Phenotype == pheno[i]], sampling = h2$samp_prev[h2$Phenotype == pheno[i]])
avengeme_res_ldsc<-rbind(avengeme_res_ldsc, data.frame(pT=pT_i,
pi0=pi0,
AUC=tmp$AUC))
}
}
# Run AVENGEME predicted AUC using AVENGEME heritability
avengeme_res_avenge<-NULL
for(pT_i in pT){
for(pi0 in seq(0.92,0.98,0.02)){
tmp<-polygenescore(nsnp=pheno_nsnp, n=h2$N[h2$Phenotype == pheno[i]], vg1 = tmp2$vg[1], pupper = c(0, pT_i), pi0 = pi0, nested = TRUE, weighted = TRUE, binary = T, prevalence = h2$pop_prev[h2$Phenotype == pheno[i]], sampling = h2$samp_prev[h2$Phenotype == pheno[i]])
avengeme_res_avenge<-rbind(avengeme_res_avenge, data.frame(pT=pT_i,
pi0=pi0,
AUC=tmp$AUC))
}
}
prs_auc_pt<-data.frame(pT=pheno_res_pTclump$pT,
AUC=pheno_res_pTclump$AUC)
prs_auc_pt1<-assoc[grepl(paste0(gwas[i],'_1$'), assoc$Predictor),]$AUC
DBSLMM_auc<-assoc[grepl('DBSLMM', assoc$Predictor),]$AUC
obs_res<-data.frame(Method=c('DBSLMM'),
AUC=c(DBSLMM_auc))
y_limit<-max(c(avengeme_res_ldsc$AUC, avengeme_res_avenge$AUC))
AVENGEME_plots[[paste0(i,'_LDSC')]]<-ggplot(avengeme_res_ldsc, aes(x=factor(pT), y=AUC, group=factor(pi0), colour=factor(pi0))) +
geom_point() +
geom_line() +
geom_point(data=prs_auc_pt, aes(x=factor(pT), y=AUC), colour='black', shape=15) +
geom_line(data=prs_auc_pt, aes(x=factor(pT), y=AUC), colour='black') +
theme_cowplot(12) +
ylim(0.5,y_limit) +
labs(x='pT', y='AUC', title=paste0(pheno[i],': LDSC h2'), colour='pi0') +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)) +
geom_hline(data=obs_res, aes(yintercept=AUC))
AVENGEME_plots[[paste0(i,'_AVENG')]]<-ggplot(avengeme_res_avenge, aes(x=factor(pT), y=AUC, group=factor(pi0),colour=factor(pi0))) +
geom_point() +
geom_line() +
geom_point(data=prs_auc_pt, aes(x=factor(pT), y=AUC), colour='black', shape=15) +
geom_line(data=prs_auc_pt, aes(x=factor(pT), y=AUC), colour='black') +
theme_cowplot(12) +
ylim(0.5,y_limit) +
labs(x='pT', y='AUC', title=paste0(pheno[i],': AVENGEME h2'), colour='pi0') +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)) +
geom_hline(data=obs_res, aes(yintercept=AUC))
avengeme_res_ldsc_best_94<-max(avengeme_res_ldsc$AUC[as.character(avengeme_res_ldsc$pi0) == 0.94])
avengeme_res_avenge_best_94<-max(avengeme_res_avenge$AUC[as.character(avengeme_res_avenge$pi0) == 0.94])
AUC_pred<-rbind(AUC_pred, data.frame(Phenotype=pheno[i],
N=h2$N[h2$Phenotype == pheno[i]],
AVENGEME_vg1=tmp2$vg[1],
AVENGEME_pi0=tmp2$pi0[1],
vg1=h2$h2[h2$Phenotype == pheno[i]],
Intercept=h2$int[h2$Phenotype == pheno[i]],
prevalence=h2$pop_prev[h2$Phenotype == pheno[i]],
sampling=h2$samp_prev[h2$Phenotype == pheno[i]],
NSNP=pheno_nsnp,
pred_AUC_LDSC=avengeme_res_ldsc_best_94,
pred_AUC_AVENG=avengeme_res_avenge_best_94,
pTclump_pt1_AUC=prs_auc_pt1,
DBSLMM_AUC=DBSLMM_auc))
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/AVENGEME_AUC.png'), units='px', res=300, width=2200, height=6000)
plot_grid(plotlist=AVENGEME_plots, ncol = 2)
dev.off()
AUC_pred$Est_Obs_diff_LDSC<-AUC_pred$pred_AUC_LDSC-AUC_pred$DBSLMM_AUC
write.csv(AUC_pred, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/AVENGEME_AUC.csv', row.names=F, quote = F)
# The LDSC intercept is below 1 for some GWAS leading to inaccurate h2 estimates
# When using AVENGEME estimates of h2, the optimal pT assuming a p0 of 0.94 is fairly close to the observed DBSLMM AUC. Its accuracy depends on how close the observed pi0 of these outcomes is to 0.94.
# Integration of polygenicity estimates based on GWAS summary statistics would be a future direction.
###########
# Estimate for continuous outcomes
###########
library(avengeme)
library(ggplot2)
library(cowplot)
pheno=c('Intelligence','BMI','Height')
gwas=c('COLL01','BODY04','HEIG03')
h2<-fread('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/ldsc_h2_obs.csv')
R2_pred<-NULL
AVENGEME_plots<-list()
for(i in 1:length(pheno)){
# Read in observed R2 estimates
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',pheno[i],'/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
# Estimate heritability using AVENGEME
pheno_nsnp<-NULL
tmp<-fread(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/pt_clump/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.NSNP_per_pT'))
pheno_nsnp<-tmp$NSNP[length(tmp$NSNP)]
pheno_res_pTclump<-assoc[grepl('PredFile1',assoc$Predictor),]
pT<-as.numeric(gsub('e.','e-',gsub('.*_','',pheno_res_pTclump$Predictor)))
pheno_res_pTclump$pT<-pT
tmp2<-estimatePolygenicModel(p=pheno_res_pTclump$BETA/pheno_res_pTclump$SE,
nsnp=pheno_nsnp,
n=c(h2$N[h2$Phenotype == pheno[i]], pheno_res_pTclump$N[1]),
pupper = c(0,pT),
binary = F,
fixvg2pi02 = T,
alpha = 0.05)
# Run AVENGEME predicted R2 using LDSC heritability
avengeme_res_ldsc<-NULL
for(pT_i in pT){
for(pi0 in seq(0.92,0.98,0.02)){
tmp<-polygenescore(nsnp=pheno_nsnp, n=h2$N[h2$Phenotype == pheno[i]], vg1 = h2$h2[h2$Phenotype == pheno[i]], pupper = c(0, pT), pi0 = pi0, nested = TRUE, weighted = TRUE, binary = F)
avengeme_res_ldsc<-rbind(avengeme_res_ldsc, data.frame(pT=pT,
pi0=pi0,
R2=tmp$R2))
}
}
# Run AVENGEME predicted R2 using AVENGEME heritability
avengeme_res_avenge<-NULL
for(pT_i in pT){
for(pi0 in seq(0.92,0.98,0.02)){
tmp<-polygenescore(nsnp=pheno_nsnp, n=h2$N[h2$Phenotype == pheno[i]], vg1 = tmp2$vg[1], pupper = c(0, pT), pi0 = pi0, nested = TRUE, weighted = TRUE, binary = F)
avengeme_res_avenge<-rbind(avengeme_res_avenge, data.frame(pT=pT,
pi0=pi0,
R2=tmp$R2))
}
}
prs_R2_pt<-data.frame(pT=pheno_res_pTclump$pT,
R2=pheno_res_pTclump$Obs_R2)
prs_R2_pt1<-assoc[grepl(paste0(gwas[i],'_1$'), assoc$Predictor),]$Obs_R2
DBSLMM_R2<-assoc[grepl('DBSLMM', assoc$Predictor),]$Obs_R2
obs_res<-data.frame(Method='DBSLMM',
R2=DBSLMM_R2)
y_limit<-max(c(avengeme_res_ldsc$R2, avengeme_res_avenge$R2))
AVENGEME_plots[[paste0(i,'_LDSC')]]<-ggplot(avengeme_res_ldsc, aes(x=factor(pT), y=R2, group=factor(pi0),colour=factor(pi0))) +
geom_point() +
geom_line() +
geom_point(data=prs_R2_pt, aes(x=factor(pT), y=R2), colour='black', shape=15) +
geom_line(data=prs_R2_pt, aes(x=factor(pT), y=R2), colour='black') +
theme_cowplot(12) +
ylim(0,y_limit) +
labs(x='pT', y='R2', title=paste0(pheno[i],': LDSC h2'), colour='pi0') +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)) +
geom_hline(data=obs_res, aes(yintercept=R2))
AVENGEME_plots[[paste0(i,'_AVENG')]]<-ggplot(avengeme_res_avenge, aes(x=factor(pT), y=R2, group=factor(pi0),colour=factor(pi0))) +
geom_point() +
geom_line() +
geom_point(data=prs_R2_pt, aes(x=factor(pT), y=R2), colour='black', shape=15) +
geom_line(data=prs_R2_pt, aes(x=factor(pT), y=R2), colour='black') +
theme_cowplot(12) +
ylim(0,y_limit) +
labs(x='pT', y='R2', title=paste0(pheno[i],': AVENGEME h2'), colour='pi0') +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)) +
geom_hline(data=obs_res, aes(yintercept=R2))
avengeme_res_ldsc_best_94<-max(avengeme_res_ldsc$R2[as.character(avengeme_res_ldsc$pi0) == 0.94])
avengeme_res_avenge_best_94<-max(avengeme_res_avenge$R2[as.character(avengeme_res_avenge$pi0) == 0.94])
R2_pred<-rbind(R2_pred, data.frame(Phenotype=pheno[i],
N=h2$N[h2$Phenotype == pheno[i]],
AVENGEME_vg1=tmp2$vg[1],
AVENGEME_pi0=tmp2$pi0[1],
vg1=h2$h2[h2$Phenotype == pheno[i]],
Intercept=h2$int[h2$Phenotype == pheno[i]],
NSNP=pheno_nsnp,
pred_R2_LDSC=avengeme_res_ldsc_best_94,
pred_R2_AVENG=avengeme_res_avenge_best_94,
pTclump_pt1_R2=prs_R2_pt1,
DBSLMM_R2=DBSLMM_R2))
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/AVENGEME_R2.png'), units='px', res=300, width=2200, height=2750)
plot_grid(plotlist=AVENGEME_plots, ncol = 2)
dev.off()
R2_pred$Est_Obs_diff_LDSC<-R2_pred$pred_R2_LDSC-R2_pred$DBSLMM_R2
write.csv(R2_pred, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/AVENGEME_R2.csv', row.names=F, quote = F)
# Note: I considered using GCTB to estimate heritability and polygenicity for AVENGEME. The estimates are often concordant, but heritability and polygenicity estimates do vary.Show results
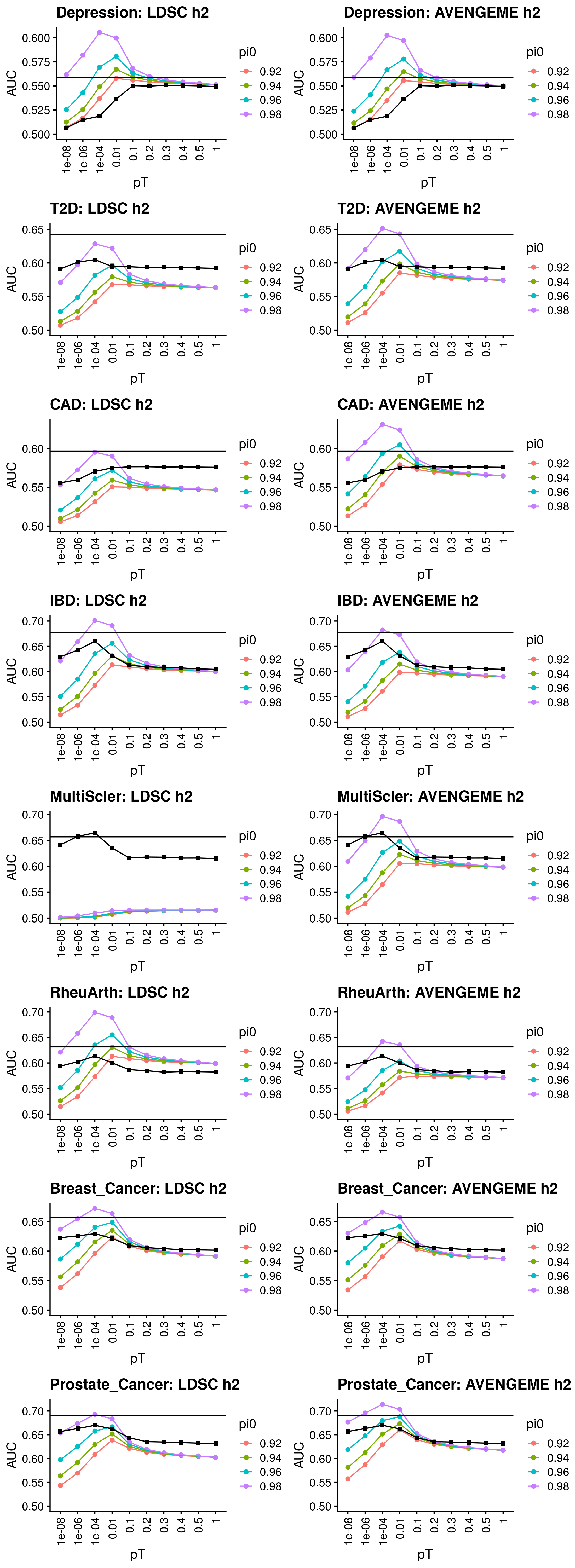
| Phenotype | N | AVENGEME h2 | LDSC h2 | LDSC intercept | Prevalence | Sampling | Est. AUC (LDSC h2) | Est. AUC (AVENGEME h2) | Obs. AUC (pT1) | Obs. AUC (DBSLMM) |
|---|---|---|---|---|---|---|---|---|---|---|
| Depression | 143265 | 0.097 | 0.100 | 0.997 | 0.150 | 0.318 | 0.567 | 0.565 | 0.549 | 0.559 |
| T2D | 152599 | 0.140 | 0.118 | 0.999 | 0.050 | 0.168 | 0.580 | 0.599 | 0.592 | 0.642 |
| CAD | 184305 | 0.078 | 0.055 | 0.887 | 0.030 | 0.330 | 0.559 | 0.590 | 0.576 | 0.597 |
| IBD | 34652 | 0.165 | 0.183 | 1.069 | 0.013 | 0.592 | 0.631 | 0.615 | 0.604 | 0.677 |
| MultiScler | 27148 | 0.133 | 0.020 | 1.062 | 0.002 | 0.360 | 0.515 | 0.623 | 0.615 | 0.657 |
| RheuArth | 58284 | 0.101 | 0.143 | 1.065 | 0.005 | 0.246 | 0.631 | 0.584 | 0.582 | 0.632 |
| Breast_Cancer | 228951 | 0.142 | 0.150 | 1.103 | 0.125 | 0.537 | 0.635 | 0.629 | 0.602 | 0.658 |
| Prostate_Cancer | 140254 | 0.218 | 0.188 | 1.081 | 0.125 | 0.564 | 0.652 | 0.673 | 0.632 | 0.691 |
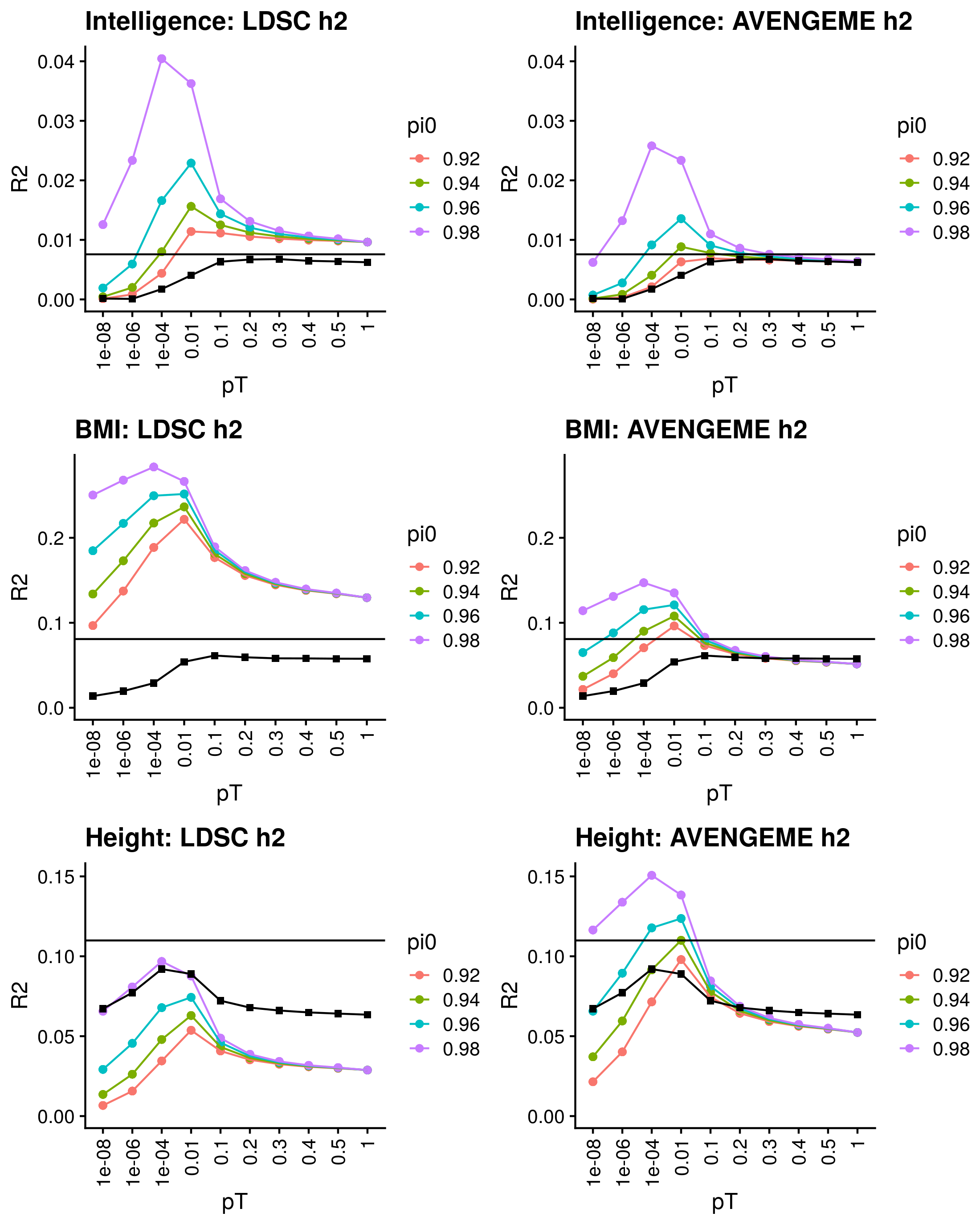
Estimates of R2 using AVENGEME/LDSC
| Phenotype | N | AVENGEME h2 | LDSC h2 | LDSC intercept | Est. R2 (LDSC h2) | Est. R2 (AVENGEME h2) | Obs. R2 (pT1) | Obs. R2 (DBSLMM) |
|---|---|---|---|---|---|---|---|---|
| Intelligence | 95427 | 0.085 | 0.105 | 1.022 | 0.016 | 0.009 | 0.006 | 0.008 |
| BMI | 252064 | 0.178 | 0.312 | 1.328 | 0.236 | 0.108 | 0.058 | 0.081 |
| Height | 233681 | 0.183 | 0.130 | 0.672 | 0.063 | 0.110 | 0.063 | 0.110 |
2.1.2 GWIZ
We will calculate the AUC using GWIZ using the leave one out meta-analysis summary statistics, and then average the AUC results. This will hopefully tell us how much variance GWAS expects the LOO sumstats to explain in the left out subset.
2.1.2.1 Format GWAS summary statistics
Show code
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
samp_prev=c(0.318,0.168,0.33,0.592,0.36,0.246,0.537,0.564)
dir.create('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/GWIZ')
for(i in 1:length(pheno)){
# Read in GWAS sumstats
sumstats<-fread(paste0(gwas_rep_qcd,'/',gwas[i],'.cleaned.gz'))
sumstats$phenotype<-'n/a'
sumstats$dataset<-'n/a'
sumstats$model<-'additive'
if(sum(names(sumstats) == 'Ncas') == 0){
sumstats$Ncas<-round(sumstats$N*samp_prev[i])
sumstats$Ncon<-round(sumstats$N*(1-samp_prev[i]))
}
sumstats$control_size<-sumstats$Ncon
sumstats$case_size<-sumstats$Ncas
if(sum(names(sumstats) == 'REF.FREQ') > 0){
sumstats$FREQ<-sumstats$REF.FREQ
sumstats$REF.FREQ<-NULL
}
if(sum(names(sumstats) == 'OR') == 0){
sumstats$OR<-exp(sumstats$BETA)
}
# OR and MAFs to all be corresponding to the risk allele
sumstats$FREQ[sumstats$OR < 1]<-1-sumstats$FREQ[sumstats$OR < 1]
names(sumstats)[names(sumstats) == 'FREQ']<-'risk_allele_freq'
sumstats$BETA<-log(sumstats$OR)
sumstats$OR[sumstats$OR < 1]<-exp(-sumstats$BETA[sumstats$OR < 1])
# Extract LD independent variants
ld_indep<-NULL
for(chr in 1:22){
ld_indep<-rbind(ld_indep, fread(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/pt_clump/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.chr',chr,'.range_values')))
}
# Filter genome-wide variants
ld_indep_sig<-ld_indep[ld_indep$V2 < 1e-8]
sumstats<-sumstats[(sumstats$SNP %in% ld_indep_sig$V1),]
sumstats<-sumstats[,c('phenotype','dataset','SNP','control_size','case_size','risk_allele_freq','OR','model'),with=F]
names(sumstats)[names(sumstats) == 'SNP']<-'accession'
sumstats$model<-rep(c('recessive','dominant'),dim(sumstats)[1])[1:dim(sumstats)[1]]
fwrite(sumstats, paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/GWIZ/',gwas[i],'.GWIZ.csv'), sep=',', na='NA', quote=F)
}2.1.2.2 Run GWIZ
GWIZ is an R script. It needs to modification: - It doesn’t allow input files, software and output to be in different directories - It uses the png function which does’t work on Rosalind - It prints the contents of the GWAS sumstats repeatedly and GWIZ is an Rscript that must be modified to allow for different input and output files. I have edited the script slightly so all files don’t have be stored in the same directory, and can be easily run in parallel.
Show code
# Run GWIZ for each phenotype and subset
for gwas in $(echo DEPR07 DIAB05 COAD01 INFB01 SCLE03 RHEU02 BRCA01 PRCA01); do
sbatch -p brc,shared --mem 10G /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/gwizer/gwizer.R \
--sumstats /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/GWIZ/${gwas}.GWIZ.csv \
--gwiz /mnt/lustre/users/k1806347/Software/GWIZ-Rscript-master \
--output /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/GWIZ/${gwas}/res_${gwas}
done2.1.2.3 Compare observed and estimate AUC
Show code
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
AUC_comp<-NULL
for(i in 1:length(pheno)){
GWIZ_AUC_i<-NULL
GWIZ_AUC_i<-read.table(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/GWIZ/',gwas[i],'/res_',gwas[i],'.log'), sep='&', header=F)$V1
GWIZ_AUC_i_auc<-as.numeric(gsub('AUC = ','',GWIZ_AUC_i[grepl('AUC = ', GWIZ_AUC_i)]))
pheno_res_pTclump<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',pheno[i],'/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
Obs_AUC<-pheno_res_pTclump$AUC[grepl('pT.clump', pheno_res_pTclump$Predictor) & grepl('1e.06', pheno_res_pTclump$Predictor)]
AUC_comp<-rbind(AUC_comp, data.frame(Phenotype=pheno[i],
Obs_AUC=Obs_AUC,
GWIZ_AUC=GWIZ_AUC_i_auc))
}
write.csv(AUC_comp, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/GWIZ_AUC.csv', row.names=F, quote=F)
# GWIZ is not performing well, except for T2D, CAD, IBD, and Depression. Again, I think this might be due to improper preparation of GWAS summary statistics. GWIZ is also running quite slow, and would need to be run GW to get accurate estimates for polygenic outcomes. I don't think this is a viable option. This method is also not suitable for continous outcomes.Show results
| Phenotype | Obs_AUC | GWIZ_AUC |
|---|---|---|
| Depression | 0.5149628 | 0.5046247 |
| T2D | 0.6011381 | 0.5715527 |
| CAD | 0.5598304 | 0.5610959 |
| IBD | 0.6424438 | 0.6745935 |
| MultiScler | 0.6577221 | 0.5523625 |
| RheuArth | 0.6024688 | 0.7521208 |
| Breast_Cancer | 0.6258635 | 0.7565655 |
| Prostate_Cancer | 0.6631618 | 0.8004651 |
2.1.3 Lassosum
Lassosum has a pseudovalidate option which estimates the correlation between predicted and observed values across shrinkage parameters. Although, its ability to estimate the best shrinakge parameter, the estimated correlation for the top shrinkage parameter appears similar to the observed correlation. We can convert the correlation into an R2 for continuous outcomes, and possible into an AUC, but converting to observed R2 into a liability R2, and then into an AUC. This would also be more suitable to predicting R2/AUC for the more modern shrinkage PRS methods, since lassosum is one.
2.1.3.1 Pseudovalidate
Show code
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
##########
# Run excluding MHC
##########
# Create directory
mkdir /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum
# Create file listing GWAS that haven't been processed.
> /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/todo_noMHC.txt
for gwas in $(echo DEPR07 COLL01 BODY04 HEIG03 DIAB05 COAD01 INFB01 SCLE03 RHEU02 BRCA01 PRCA01);do
if [ ! -f /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/${gwas}_noMHC/lassosum_pseudo_${gwas}.pseudovalidate.png ]; then
echo $gwas >> /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/todo_noMHC.txt
fi
done
# Create shell script to run using sbatch
cat > /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/sbatch_noMHC.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem=10G
#SBATCH -n 1
#SBATCH -J lassosum
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(sed "${SLURM_ARRAY_TASK_ID}q;d" /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/todo_noMHC.txt)
echo ${gwas}
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/lassosum_pseudovalidate/lassosum_pseudovalidate.R \
--ref_plink_gw ${Geno_1KG_dir}/1KGPhase3.w_hm3.GW \
--ref_keep ${Geno_1KG_dir}/keep_files/EUR_samples.keep \
--sumstats ${gwas_rep_qcd}/${gwas}.cleaned.gz \
--prune_mhc T \
--output /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/${gwas}_noMHC/lassosum_pseudo_${gwas} \
--plink /users/k1806347/brc_scratch/Software/plink1.9.sh \
--n_cores 1
EOF
sbatch --array 1-$(wc -l /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/todo_noMHC.txt | cut -d' ' -f1)%5 /scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/sbatch_noMHC.sh
2.1.3.2 Compare observed and estimate AUC
Show code
library(data.table)
#####
# Binary outcomes
#####
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
samp_prev=c(0.318,0.168,0.33,0.592,0.36,0.246,0.537,0.564,0.318)
AUC_comp<-NULL
for(i in 1:length(pheno)){
lasso_AUC_i<-NULL
lasso_AUC_i<-read.table(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/',gwas[i],'_noMHC/lassosum_pseudo_',gwas[i],'.log'), sep='&', header=F)$V1
lasso_AUC_i_r<-as.numeric(gsub('value = ','',lasso_AUC_i[grepl('value = ', lasso_AUC_i)]))
n_case<-samp_prev[i]
n_con<-1-samp_prev[i]
a<-(n_case+n_con)^2/(n_case*n_con)
lasso_AUC_i_d<-sqrt(a)*lasso_AUC_i_r/sqrt(1-lasso_AUC_i_r^2)
lasso_AUC_i_auc <- pnorm(lasso_AUC_i_d/sqrt(2), 0, 1)
pheno_res<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',pheno[i],'/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
Obs_pT_clump_AUC<-pheno_res$AUC[grepl('_1$', pheno_res$Predictor)]
Obs_DBSLMM_AUC<-pheno_res$AUC[grepl('_DBSLMM', pheno_res$Predictor)]
AUC_comp<-rbind(AUC_comp, data.frame(Phenotype=pheno[i],
Obs_pTclump_pt1_AUC=Obs_pT_clump_AUC,
Obs_DBSLMM_AUC=Obs_DBSLMM_AUC,
Est_lasso_AUC=lasso_AUC_i_auc))
}
AUC_comp$Est_Obs_diff_DBSLMM<-AUC_comp$Est_lasso_AUC-AUC_comp$Obs_DBSLMM_AUC
write.csv(AUC_comp, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/lassosum_AUC.csv', row.names=F, quote=F)
#####
# Continuous outcomes
#####
pheno=c('Intelligence','BMI','Height')
gwas=c('COLL01','BODY04','HEIG03')
R2_comp<-NULL
for(i in 1:length(pheno)){
lasso_R2_i<-NULL
lasso_R2_i<-read.table(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/',gwas[i],'_noMHC/lassosum_pseudo_',gwas[i],'.log'), sep='&', header=F)$V1
lasso_R2_i_r<-as.numeric(gsub('value = ','',lasso_R2_i[grepl('value = ', lasso_R2_i)]))
lasso_R2_i_r2<-lasso_R2_i_r^2
pheno_res<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',pheno[i],'/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
Obs_pT_clump_R2<-pheno_res$Obs_R2[grepl('_1$', pheno_res$Predictor)]
Obs_DBSLMM_R2<-pheno_res$Obs_R2[grepl('_DBSLMM', pheno_res$Predictor)]
R2_comp<-rbind(R2_comp, data.frame(Phenotype=pheno[i],
Obs_pTclump_pT1_R2=Obs_pT_clump_R2,
Obs_DBSLMM_R2=Obs_DBSLMM_R2,
lasso_R2=lasso_R2_i_r2))
}
R2_comp$Est_Obs_diff_DBSLMM<-R2_comp$lasso_R2-R2_comp$Obs_DBSLMM_R2
write.csv(R2_comp, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/lassosum_R2.csv', row.names=F, quote=F)
# This is looking really promising. Lassosum has some trouble with IBD, MultiScler and RheuArth, but otherwise is very accurate. These outcomes are all rarer and autoimmune disorders. I have run without mhc region but results still poor for these outcomes. Discordant findings may be due to phenotype heterogeneity between GWAS and UKB.Show results
| Phenotype | Observed AUC | Estimated AUC | Difference |
|---|---|---|---|
| Depression | 0.559 | 0.555 | -0.004 |
| T2D | 0.642 | 0.651 | 0.010 |
| CAD | 0.597 | 0.624 | 0.027 |
| IBD | 0.677 | 0.792 | 0.115 |
| MultiScler | 0.657 | 0.785 | 0.128 |
| RheuArth | 0.632 | 0.500 | -0.132 |
| Breast_Cancer | 0.658 | 0.698 | 0.040 |
| Prostate_Cancer | 0.691 | 0.721 | 0.030 |
| Phenotype | Observed R2 | Estimated R2 | Difference |
|---|---|---|---|
| Intelligence | 0.008 | 0.023 | 0.015 |
| BMI | 0.081 | 0.074 | -0.007 |
| Height | 0.110 | 0.156 | 0.046 |
Based on these results, none of these approaches are brilliant. The AVENGEME/LDSC method is limited by the accuracy of the LDSC heritability estimate, and the ‘medium’ polygencity assumed by using a pi0 of 0.94. The lassosum approach does fairly well, but is not good for the autoimmune disorders. I think we should use the lassosum estimate unless it is R2=0 or AUC=0.5.
2.1.4 Compare absolute estimates/values when using AVENGEME AUC/R2
2.1.4.1 Binary
Show code
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
ccprobs.f <- function(PRS_auc=0.641, prev=0.7463, n_quantile=20){
# Convert AUC into cohen's d
d <- sqrt(2)*qnorm(PRS_auc)
# Set mean difference between cases and control polygenic scores
mu_case <- d
mu_control <- 0
# Estimate mean and variance of polygenic scores across case and control
varPRS <- prev*(1+(d^2) - (d*prev)^2) + (1-prev)*(1 - (d*prev)^2)
E_PRS <- d*prev
# Estimate polygenic score quantiles
by_quant<-1/n_quantile
p_quant <- seq(by_quant, 1-by_quant, by=by_quant)
quant_vals_PRS <- rep(0, length(p_quant))
quant_f_solve <- function(x, prev, d, pq){prev*pnorm(x-d) + (1-prev)*pnorm(x) - pq}
for(i in 1:length(p_quant)){
quant_vals_PRS[i] <- unlist(uniroot(quant_f_solve, prev=prev, d=d, pq= p_quant[i], interval=c(-2.5, 2.5), extendInt = "yes", tol=6e-12)$root)
}
# Create a table for output
ul_qv_PRS <- matrix(0, ncol=2, nrow=n_quantile)
ul_qv_PRS[1,1] <- -Inf
ul_qv_PRS[2:n_quantile,1] <- quant_vals_PRS
ul_qv_PRS[1:(n_quantile-1),2] <- quant_vals_PRS
ul_qv_PRS[n_quantile,2] <- Inf
ul_qv_PRS<-cbind(ul_qv_PRS, (ul_qv_PRS[,1:2]-E_PRS)/sqrt(varPRS))
# Estimate case control proportion for each quantile
prob_quantile_case <- pnorm(ul_qv_PRS[,2], mean = mu_case) - pnorm(ul_qv_PRS[,1], mean = mu_case)
prob_quantile_control <- pnorm(ul_qv_PRS[,2], mean = mu_control) - pnorm(ul_qv_PRS[,1], mean = mu_control)
p_case_quantile <- (prob_quantile_case*prev)/by_quant
p_cont_quantile <- (prob_quantile_control*(1-prev))/by_quant
# Estimate OR comparing each quantile to bottom quantile
OR <- p_case_quantile/p_cont_quantile
OR <- OR/OR[1]
# Return output
out <- cbind(ul_qv_PRS[,3:4],p_cont_quantile, p_case_quantile, OR)
row.names(out) <- 1:n_quantile
colnames(out) <- c("q_min", "q_max","p_control", "p_case", "OR")
data.frame(out)
}
# Read in AVENGEME AUC
AVENGEME_AUC<-read.csv('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/AVENGEME_AUC.csv')
# Run analysis for each phenotype
res_all<-NULL
cor_res<-NULL
plots_all<-list()
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/DBSLMM/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.DBSLMM_profiles'))
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_DBSLMM')), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
# Read in AUC for PRS
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',files$pheno[i],'/UKBB.w_hm3.',files$gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
prs_auc<-assoc[grepl('DBSLMM', assoc$Predictor),]$AUC
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs$obs_quant<-as.numeric(cut( pheno_prs$prs, obs_quant, include.lowest = T))
# Calculate proportion of each quantile that are cases
obs_cc<-NULL
for(k in 1:n_quant){
obs_cc<-rbind(obs_cc, data.frame(Phenotype=files$pheno[i],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
p_control=1-mean(pheno_prs$pheno[pheno_prs$obs_quant == k]),
p_case=mean(pheno_prs$pheno[pheno_prs$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_cc<-ccprobs.f(PRS_auc = prs_auc, prev=mean(pheno_prs$pheno), n_quantile = n_quant)
est_cc$OR<-NULL
est_cc<-data.frame(Phenotype=files$pheno[i],Type='Estimated',Quantile=1:n_quant, est_cc)
est_quant<-sort(unique(c(est_cc$q_min, est_cc$q_max)))
pheno_prs$est_quant<-as.numeric(cut( pheno_prs$prs, est_quant, include.lowest = T))
tmp<-cor.test(obs_cc$p_case, est_cc$p_case)
tmp2<-abs(est_cc$p_case-obs_cc$p_case)/obs_cc$p_case
# Assign individuals to estimated PRS quantiles
AVENGEME_est_cc<-ccprobs.f(PRS_auc = AVENGEME_AUC$pred_AUC_LDSC[AVENGEME_AUC$Phenotype == files$pheno[i]], prev=mean(pheno_prs$pheno), n_quantile = n_quant)
AVENGEME_est_cc$OR<-NULL
AVENGEME_est_cc<-data.frame(Phenotype=files$pheno[i],Type="Estimated\n(AVENGEME)",Quantile=1:n_quant, AVENGEME_est_cc)
AVENGEME_est_quant<-sort(unique(c(AVENGEME_est_cc$q_min, AVENGEME_est_cc$q_max)))
pheno_prs$AVENGEME_est_quant<-as.numeric(cut( pheno_prs$prs, AVENGEME_est_quant, include.lowest = T))
tmp3<-cor.test(obs_cc$p_case, AVENGEME_est_cc$p_case)
tmp4<-abs(AVENGEME_est_cc$p_case-obs_cc$p_case)/obs_cc$p_case
# Estimate correlation between observed and expected
cor_res<-rbind(cor_res,data.frame(Phenotype=files$pheno[i],
Cor=tmp$estimate,
Low95CI=tmp$conf.int[1],
High95CI=tmp$conf.int[2],
Mean_perc_diff=mean(tmp2),
Cor_AVENGEME=tmp3$estimate,
Low95CI_AVENGEME=tmp3$conf.int[1],
High95CI_AVENGEME=tmp3$conf.int[2],
Mean_perc_diff_AVENGEME=mean(tmp4),
N=length(pheno_prs$pheno),
Ncas=sum(pheno_prs$pheno == 1),
Ncon=sum(pheno_prs$pheno == 0)))
quant_comp<-do.call(rbind,list(obs_cc, est_cc, AVENGEME_est_cc))
res_all<-rbind(res_all, quant_comp)
library(ggplot2)
library(cowplot)
plots_all[[i]]<-ggplot(quant_comp, aes(x=Quantile, y=p_case, colour=Type)) +
geom_point(alpha=0.8) +
geom_line(alpha=0.8) +
labs(x='PRS quantile', y=paste0('Proportion with ',files$pheno[i]), title=files$pheno[i], colour='Method') +
theme_cowplot(12)
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PropCC_Comp_AVENGEME.png'), units='px', res=300, width=2000, height=3000)
plot_grid(plotlist=plots_all, ncol = 2)
dev.off()
write.csv(cor_res, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PropCC_Comp_AVENGEME.csv', row.names=F, quote=F)
ex_est<-res_all[res_all$Phenotype == 'MultiScler' & res_all$Type == "Estimated\n(AVENGEME)" & res_all$Quantile == 20,]$p_case
ex_obs<-res_all[res_all$Phenotype == 'MultiScler' & res_all$Type == 'Observed' & res_all$Quantile == 20,]$p_case
ex_est/ex_obs
ex_est<-res_all[res_all$Phenotype == 'IBD' & res_all$Type == "Estimated\n(AVENGEME)" & res_all$Quantile == 20,]$p_case
ex_obs<-res_all[res_all$Phenotype == 'IBD' & res_all$Type == 'Observed' & res_all$Quantile == 20,]$p_case
ex_est/ex_obs
ex_est<-res_all[res_all$Phenotype == 'T2D' & res_all$Type == "Estimated\n(AVENGEME)" & res_all$Quantile == 20,]$p_case
ex_obs<-res_all[res_all$Phenotype == 'T2D' & res_all$Type == 'Observed' & res_all$Quantile == 20,]$p_case
ex_est/ex_obsShow results
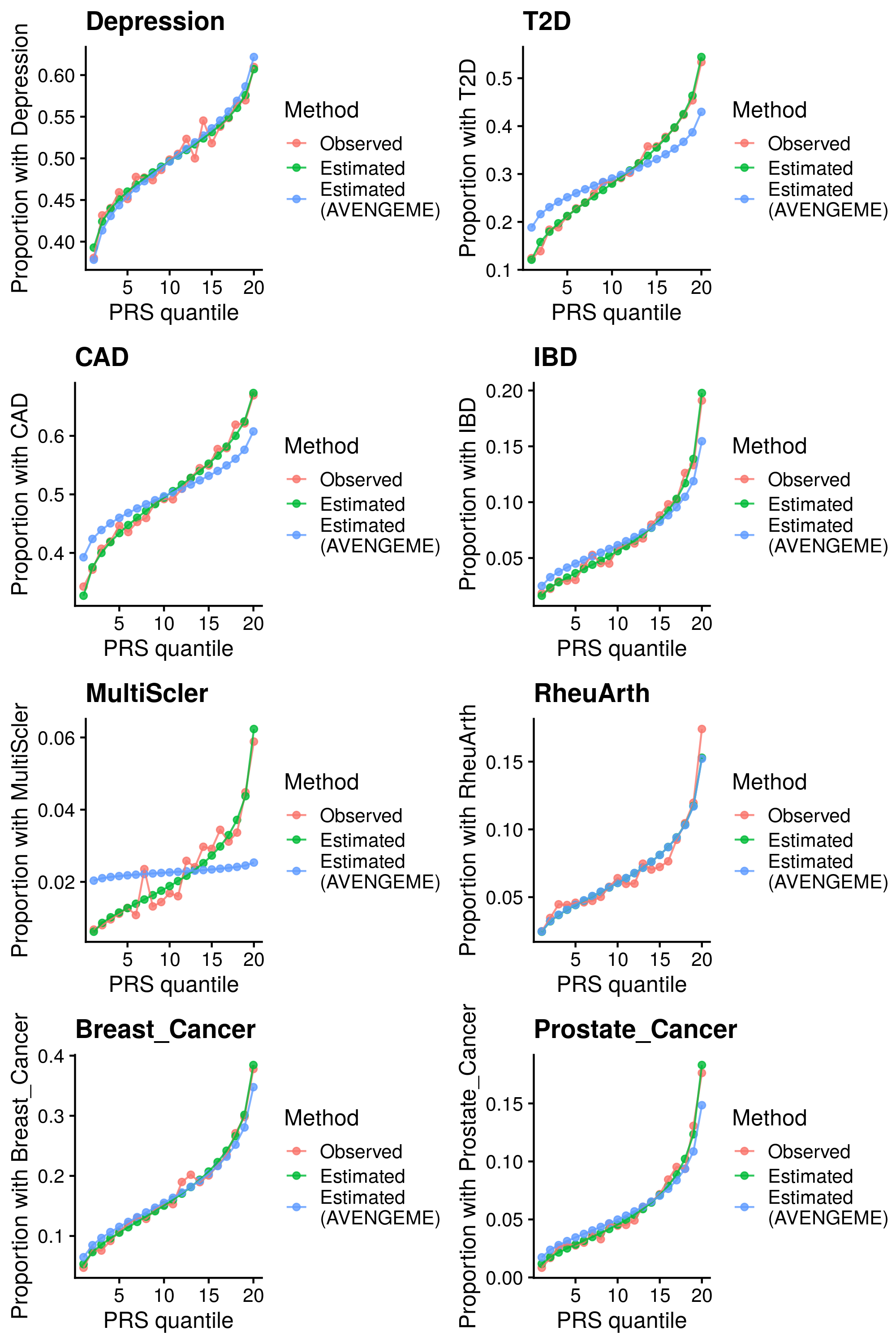
| Phenotype | Cor | Low95CI | High95CI | Mean_perc_diff | Cor_AVENGEME | Low95CI_AVENGEME | High95CI_AVENGEME | Mean_perc_diff_AVENGEME | N | Ncas | Ncon |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Depression | 0.9845782 | 0.9605776 | 0.9940117 | 0.0150485 | 0.9844983 | 0.9603757 | 0.9939806 | 0.0193074 | 49999 | 24999 | 25000 |
| T2D | 0.9968126 | 0.9917731 | 0.9987670 | 0.0236348 | 0.9968601 | 0.9918954 | 0.9987854 | 0.1530956 | 49999 | 14888 | 35111 |
| CAD | 0.9943181 | 0.9853636 | 0.9978003 | 0.0153319 | 0.9938163 | 0.9840773 | 0.9976057 | 0.0578957 | 49999 | 25000 | 24999 |
| IBD | 0.9941885 | 0.9850314 | 0.9977501 | 0.0683969 | 0.9928430 | 0.9815852 | 0.9972280 | 0.1825526 | 49999 | 3461 | 46538 |
| MultiScler | 0.9691668 | 0.9221227 | 0.9879704 | 0.1262171 | 0.9375296 | 0.8459902 | 0.9753859 | 0.6005559 | 49999 | 1137 | 48862 |
| RheuArth | 0.9813043 | 0.9523309 | 0.9927332 | 0.0684472 | 0.9810557 | 0.9517064 | 0.9926360 | 0.0678404 | 49999 | 3408 | 46591 |
| Breast_Cancer | 0.9949234 | 0.9869165 | 0.9980350 | 0.0456799 | 0.9955383 | 0.9884957 | 0.9982734 | 0.0838627 | 49999 | 8512 | 41487 |
| Prostate_Cancer | 0.9939174 | 0.9843364 | 0.9976449 | 0.0870004 | 0.9930048 | 0.9819994 | 0.9972908 | 0.1788078 | 50000 | 2927 | 47073 |
Median Cor. = 0.9929239; Mean Cor. = 0.9843933; Min. Cor. = 0.9375296; Max. Cor. = 0.9968601
Excluding RheuArth: Median Cor. = 0.9930048; Mean Cor. = 0.98487; Min. Cor. = 0.9375296; Max. Cor. = 0.9968601
2.1.4.2 Continuous
Show code
library(data.table)
library(e1071)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
gwas<-c('COLL01','HEIG03','BODY04')
pheno<-c('Intelligence','Height','BMI')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
mean_sd_quant.f <- function(PRS_R2=0.641, Outcome_mean=1, Outcome_sd=1, n_quantile=20){
### PRS quantiles with a continuous phenotype (Y)
library(tmvtnorm)
###
E_PRS = 0
SD_PRS = sqrt(1)
E_phenotype = Outcome_mean
SD_phenotype = Outcome_sd
by_quant<-1/(n_quantile)
PRS_quantile_bounds <- qnorm(p=seq(0, 1, by=by_quant), mean= E_PRS, sd= SD_PRS)
lower_PRS_vec <- PRS_quantile_bounds[1:n_quantile]
upper_PRS_vec <- PRS_quantile_bounds[2:(n_quantile+1)]
mean_vec <- c(E_phenotype, E_PRS)
sigma_mat <- matrix(sqrt(PRS_R2)*SD_phenotype*SD_PRS, nrow=2, ncol=2)
sigma_mat[1,1] <- SD_phenotype^2
sigma_mat[2,2] <- SD_PRS^2
### mean of phenotype within the truncated PRS distribution
out_mean_Y <- rep(0, 20)
### SD of phenotype within the truncated PRS distribution
out_SD_Y <- rep(0, 20)
### cov of Y and PRS given truncation on PRS
out_cov_Y_PRS <- rep(0, 20)
### SD of PRS given truncation on PRS
out_SD_PRS <- rep(0, 20)
### mean PRS given truncation on PRS
out_mean_PRS <- rep(0, 20)
for(i in 1:n_quantile){
distribution_i <- mtmvnorm(mean = mean_vec,
sigma = sigma_mat,
lower = c(-Inf, lower_PRS_vec[i]),
upper = c(Inf, upper_PRS_vec[i]),
doComputeVariance=TRUE,
pmvnorm.algorithm=GenzBretz())
out_mean_Y[i] <- distribution_i$tmean[1]
out_mean_PRS[i] <- distribution_i$tmean[2]
out_SD_Y[i] <- sqrt(distribution_i$tvar[1,1])
out_SD_PRS[i] <- sqrt(distribution_i$tvar[2,2])
out_cov_Y_PRS[i] <- distribution_i$tvar[1,2]
}
out<-data.frame(q=1:n_quantile,
q_min=lower_PRS_vec,
q_max=upper_PRS_vec,
x_mean=out_mean_Y,
x_sd=out_SD_Y)
return(out)
out_mean_Y
out_SD_Y
out_mean_PRS
out_SD_PRS
out_cov_Y_PRS
}
# Read in AVENGEME R2
AVENGEME_R2<-read.csv('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/AVENGEME_R2.csv')
# Run analysis for each phenotype
res_all<-NULL
plots_all<-list()
cor_res<-NULL
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/DBSLMM/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.DBSLMM_profiles'))
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_DBSLMM')), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
# Read in AUC for PRS
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',files$pheno[i],'/UKBB.w_hm3.',files$gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
prs_r2<-assoc[grepl('DBSLMM', assoc$Predictor),]$Obs_R2
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs$obs_quant<-as.numeric(cut( pheno_prs$prs, obs_quant, include.lowest = T))
# Calculate mean and SD of each quantile that are cases
obs_dist<-NULL
for(k in 1:n_quant){
obs_dist<-rbind(obs_dist, data.frame(Phenotype=files$pheno[i],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
x_mean=mean(pheno_prs$pheno[pheno_prs$obs_quant == k]),
x_sd=sd(pheno_prs$pheno[pheno_prs$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_dist<-mean_sd_quant.f(PRS_R2 = prs_r2, Outcome_mean=mean(pheno_prs$pheno), Outcome_sd=sd(pheno_prs$pheno), n_quantile = n_quant)
est_dist$q<-NULL
est_dist<-data.frame(Phenotype=files$pheno[i],Type='Estimated',Quantile=1:n_quant, est_dist)
est_quant<-sort(unique(c(est_dist$q_min, est_dist$q_max)))
pheno_prs$est_quant<-as.numeric(cut( pheno_prs$prs, est_quant, include.lowest = T))
# Assign individuals to AVENGEME estimated PRS quantiles
AVENGEME_est_dist<-mean_sd_quant.f(PRS_R2 = AVENGEME_R2$pred_R2_LDSC[AVENGEME_R2$Phenotype == files$pheno[i]], Outcome_mean=mean(pheno_prs$pheno), Outcome_sd=sd(pheno_prs$pheno), n_quantile = n_quant)
AVENGEME_est_dist$q<-NULL
AVENGEME_est_dist<-data.frame(Phenotype=files$pheno[i],Type="Estimated\n(AVENGEME)",Quantile=1:n_quant, AVENGEME_est_dist)
AVENGEME_est_quant<-sort(unique(c(AVENGEME_est_dist$q_min, AVENGEME_est_dist$q_max)))
pheno_prs$AVENGEME_est_quant<-as.numeric(cut( pheno_prs$prs, AVENGEME_est_quant, include.lowest = T))
quant_comp<-do.call(rbind, list(obs_dist, est_dist, AVENGEME_est_dist))
tmp<-cor.test(obs_dist$x_mean, est_dist$x_mean)
tmp2<-cor.test(obs_dist$x_mean, AVENGEME_est_dist$x_mean)
cor_res<-rbind(cor_res,data.frame(Phenotype=files$pheno[i],
Cor_mean=tmp$estimate,
Cor_mean_Low95CI=tmp$conf.int[1],
Cor_mean_High95CI=tmp$conf.int[2],
Cor_mean_AVENGEME=tmp2$estimate,
Cor_mean_Low95CI_AVENGEME=tmp2$conf.int[1],
Cor_mean_High95CI_AVENGEME=tmp2$conf.int[2],
PercDiff_sd_mean=mean(abs(est_dist$x_sd-obs_dist$x_sd)/obs_dist$x_sd),
PercDiff_sd_mean_AVENGEME=mean(abs(AVENGEME_est_dist$x_sd-obs_dist$x_sd)/obs_dist$x_sd),
N=length(pheno_prs$pheno),
Skewness=skewness(pheno_prs$pheno)))
res_all<-rbind(res_all, quant_comp)
library(ggplot2)
library(cowplot)
plots_all[[i]]<-ggplot(quant_comp, aes(x=Quantile, y=x_mean, colour=Type)) +
geom_point(stat="identity", position=position_dodge(.9), alpha=0.8, shape=18) +
geom_errorbar(aes(ymin=x_mean-x_sd, ymax=x_mean+x_sd), width=.2, position=position_dodge(.9), alpha=0.8) +
labs(x='PRS quantile', y=paste0(pheno[i], " mean (SD)"), title=files$pheno[i], colour='Method') +
theme_cowplot(12)
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/Mean_SD_Comp_AVENGEME.png'), units='px', res=300, width=1500, height=1750)
plot_grid(plotlist=plots_all, ncol = 1)
dev.off()
write.csv(cor_res, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/Mean_SD_Comp_AVENGEME.csv', row.names=F, quote=F)Show results
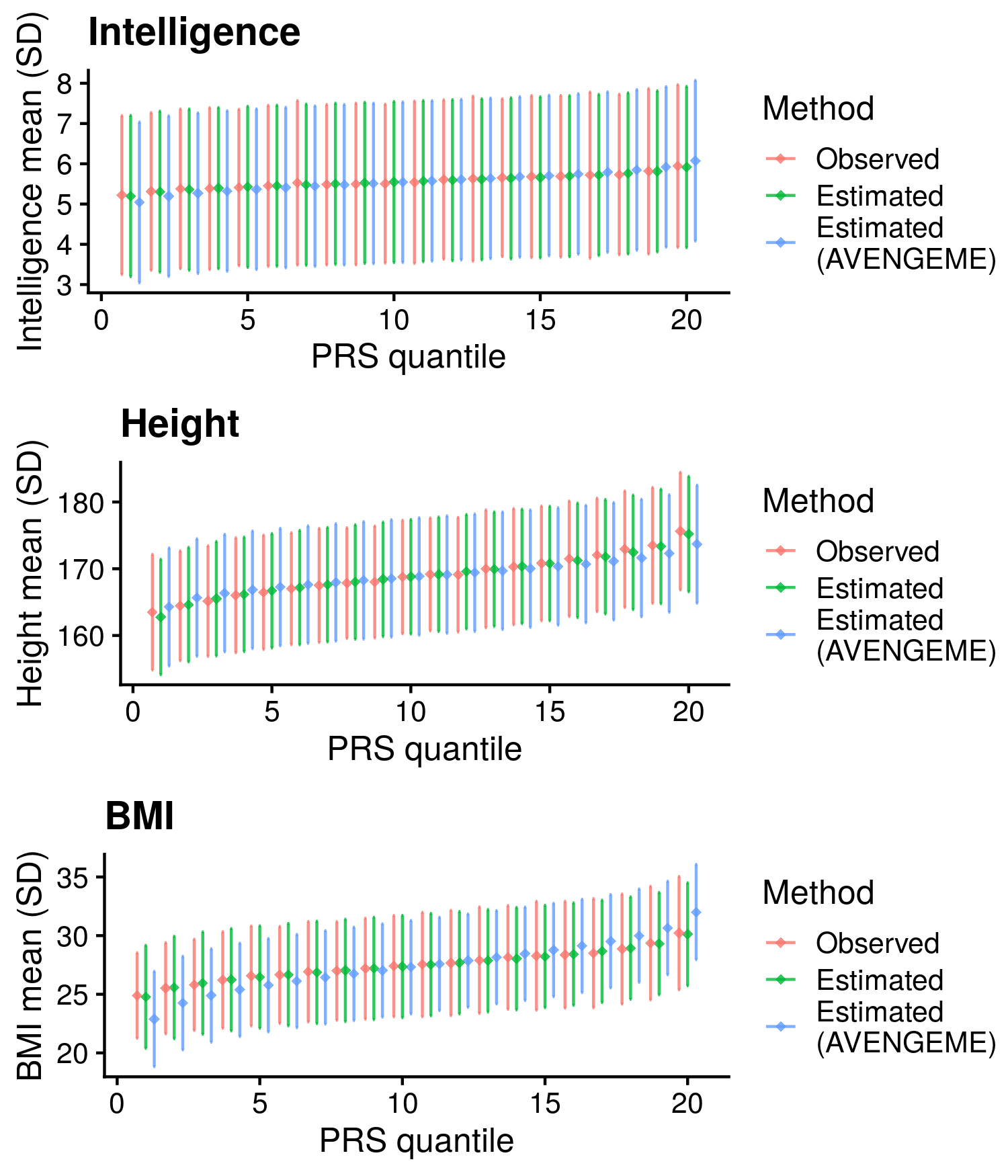
| Phenotype | Cor_mean | Cor_mean_Low95CI | Cor_mean_High95CI | Cor_mean_AVENGEME | Cor_mean_Low95CI_AVENGEME | Cor_mean_High95CI_AVENGEME | PercDiff_sd_mean | PercDiff_sd_mean_AVENGEME | N | Skewness |
|---|---|---|---|---|---|---|---|---|---|---|
| Intelligence | 0.9917264 | 0.9787310 | 0.9967944 | 0.9917264 | 0.9787310 | 0.9967944 | 0.0132982 | 0.0135699 | 50000 | 0.1440539 |
| Height | 0.9958381 | 0.9892663 | 0.9983895 | 0.9958381 | 0.9892663 | 0.9983895 | 0.0151424 | 0.0306504 | 49999 | 0.1168512 |
| BMI | 0.9981713 | 0.9952749 | 0.9992929 | 0.9981713 | 0.9952749 | 0.9992929 | 0.0596993 | 0.0959049 | 49999 | 0.5915969 |
Median Cor. of means = 0.9958381; Mean Cor. of means = 0.9952452; Min. Cor. of means = 0.9917264; Max. Cor. of means = 0.9981713
Median mean %diff of SD = 0.03065044; Mean mean %diff of SD = 0.04670842; Min. mean %diff of SD = 0.01356991; Max. mean %diff of SD = 0.09590491
2.1.5 Compare absolute estimates/values when using lassosum AUC/R2
2.1.5.1 Binary
Show code
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
pheno=c('Depression','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas=c('DEPR07','DIAB05','COAD01','INFB01','SCLE03','RHEU02','BRCA01','PRCA01')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
ccprobs.f <- function(PRS_auc=0.641, prev=0.7463, n_quantile=20){
# Convert AUC into cohen's d
d <- sqrt(2)*qnorm(PRS_auc)
# Set mean difference between cases and control polygenic scores
mu_case <- d
mu_control <- 0
# Estimate mean and variance of polygenic scores across case and control
varPRS <- prev*(1+(d^2) - (d*prev)^2) + (1-prev)*(1 - (d*prev)^2)
E_PRS <- d*prev
# Estimate polygenic score quantiles
by_quant<-1/n_quantile
p_quant <- seq(by_quant, 1-by_quant, by=by_quant)
quant_vals_PRS <- rep(0, length(p_quant))
quant_f_solve <- function(x, prev, d, pq){prev*pnorm(x-d) + (1-prev)*pnorm(x) - pq}
for(i in 1:length(p_quant)){
quant_vals_PRS[i] <- unlist(uniroot(quant_f_solve, prev=prev, d=d, pq= p_quant[i], interval=c(-2.5, 2.5), extendInt = "yes", tol=6e-12)$root)
}
# Create a table for output
ul_qv_PRS <- matrix(0, ncol=2, nrow=n_quantile)
ul_qv_PRS[1,1] <- -Inf
ul_qv_PRS[2:n_quantile,1] <- quant_vals_PRS
ul_qv_PRS[1:(n_quantile-1),2] <- quant_vals_PRS
ul_qv_PRS[n_quantile,2] <- Inf
ul_qv_PRS<-cbind(ul_qv_PRS, (ul_qv_PRS[,1:2]-E_PRS)/sqrt(varPRS))
# Estimate case control proportion for each quantile
prob_quantile_case <- pnorm(ul_qv_PRS[,2], mean = mu_case) - pnorm(ul_qv_PRS[,1], mean = mu_case)
prob_quantile_control <- pnorm(ul_qv_PRS[,2], mean = mu_control) - pnorm(ul_qv_PRS[,1], mean = mu_control)
p_case_quantile <- (prob_quantile_case*prev)/by_quant
p_cont_quantile <- (prob_quantile_control*(1-prev))/by_quant
# Estimate OR comparing each quantile to bottom quantile
OR <- p_case_quantile/p_cont_quantile
OR <- OR/OR[1]
# Return output
out <- cbind(ul_qv_PRS[,3:4],p_cont_quantile, p_case_quantile, OR)
row.names(out) <- 1:n_quantile
colnames(out) <- c("q_min", "q_max","p_control", "p_case", "OR")
data.frame(out)
}
# Read in lassosum AUC
lassosum_AUC<-read.csv('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/lassosum_AUC.csv')
# Run analysis for each phenotype
res_all<-NULL
cor_res<-NULL
plots_all<-list()
plots_all_poster<-list()
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/DBSLMM/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.DBSLMM_profiles'))
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_DBSLMM')), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
# Read in AUC for PRS
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',files$pheno[i],'/UKBB.w_hm3.',files$gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
prs_auc<-assoc[grepl('DBSLMM', assoc$Predictor),]$AUC
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs$obs_quant<-as.numeric(cut( pheno_prs$prs, obs_quant, include.lowest = T))
# Calculate proportion of each quantile that are cases
obs_cc<-NULL
for(k in 1:n_quant){
obs_cc<-rbind(obs_cc, data.frame(Phenotype=files$pheno[i],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
p_control=1-mean(pheno_prs$pheno[pheno_prs$obs_quant == k]),
p_case=mean(pheno_prs$pheno[pheno_prs$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_cc<-ccprobs.f(PRS_auc = prs_auc, prev=mean(pheno_prs$pheno), n_quantile = n_quant)
est_cc$OR<-NULL
est_cc<-data.frame(Phenotype=files$pheno[i],Type="\nEstimated\n(observed AUC)\n",Quantile=1:n_quant, est_cc)
est_quant<-sort(unique(c(est_cc$q_min, est_cc$q_max)))
pheno_prs$est_quant<-as.numeric(cut( pheno_prs$prs, est_quant, include.lowest = T))
tmp<-cor.test(obs_cc$p_case, est_cc$p_case)
tmp2<-abs(est_cc$p_case-obs_cc$p_case)/obs_cc$p_case
# Assign individuals to estimated PRS quantiles
lasso_est_cc<-ccprobs.f(PRS_auc = lassosum_AUC$Est_lasso_AUC[lassosum_AUC$Phenotype == files$pheno[i]], prev=mean(pheno_prs$pheno), n_quantile = n_quant)
lasso_est_cc$OR<-NULL
lasso_est_cc<-data.frame(Phenotype=files$pheno[i],Type="Estimated\n(lassosum AUC)\n",Quantile=1:n_quant, lasso_est_cc)
lasso_est_quant<-sort(unique(c(lasso_est_cc$q_min, lasso_est_cc$q_max)))
pheno_prs$lasso_est_quant<-as.numeric(cut( pheno_prs$prs, lasso_est_quant, include.lowest = T))
tmp3<-cor.test(obs_cc$p_case, lasso_est_cc$p_case)
tmp4<-abs(lasso_est_cc$p_case-obs_cc$p_case)/obs_cc$p_case
# Estimate correlation between observed and expected
cor_res<-rbind(cor_res,data.frame(Phenotype=files$pheno[i],
Cor=tmp$estimate,
Low95CI=tmp$conf.int[1],
High95CI=tmp$conf.int[2],
Mean_perc_diff=mean(tmp2),
Cor_lasso=tmp3$estimate,
Low95CI_lasso=tmp3$conf.int[1],
High95CI_lasso=tmp3$conf.int[2],
Mean_perc_diff_lasso=mean(tmp4),
N=length(pheno_prs$pheno),
Ncas=sum(pheno_prs$pheno == 1),
Ncon=sum(pheno_prs$pheno == 0)))
quant_comp<-do.call(rbind,list(obs_cc, est_cc, lasso_est_cc))
res_all<-rbind(res_all, quant_comp)
library(ggplot2)
library(cowplot)
plots_all[[i]]<-ggplot(quant_comp, aes(x=Quantile, y=p_case, colour=Type)) +
geom_line() +
geom_point(alpha=0.8, shape=18, size=2) +
labs(y="p(case)", title=files$pheno[i], colour='Method') +
scale_fill_manual(values=c("#208eb7", "#e3a0fa", "#284e37")) +
theme_cowplot(12)
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PropCC_Comp_lasso.png'), units='px', res=300, width=2000, height=2800)
plot_grid(plotlist=plots_all, ncol = 2)
dev.off()
write.csv(cor_res, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/PropCC_Comp_lasso.csv', row.names=F, quote=F)
ex_est<-res_all[res_all$Phenotype == 'MultiScler' & res_all$Type == "Estimated (lasso)" & res_all$Quantile == 20,]$p_case
ex_obs<-res_all[res_all$Phenotype == 'MultiScler' & res_all$Type == 'Observed' & res_all$Quantile == 20,]$p_case
ex_est/ex_obs
ex_est<-res_all[res_all$Phenotype == 'IBD' & res_all$Type == "Estimated (lasso)" & res_all$Quantile == 20,]$p_case
ex_obs<-res_all[res_all$Phenotype == 'IBD' & res_all$Type == 'Observed' & res_all$Quantile == 20,]$p_case
ex_est/ex_obs
ex_est<-res_all[res_all$Phenotype == 'T2D' & res_all$Type == "Estimated (lasso)" & res_all$Quantile == 20,]$p_case
ex_obs<-res_all[res_all$Phenotype == 'T2D' & res_all$Type == 'Observed' & res_all$Quantile == 20,]$p_case
ex_est/ex_obsShow results
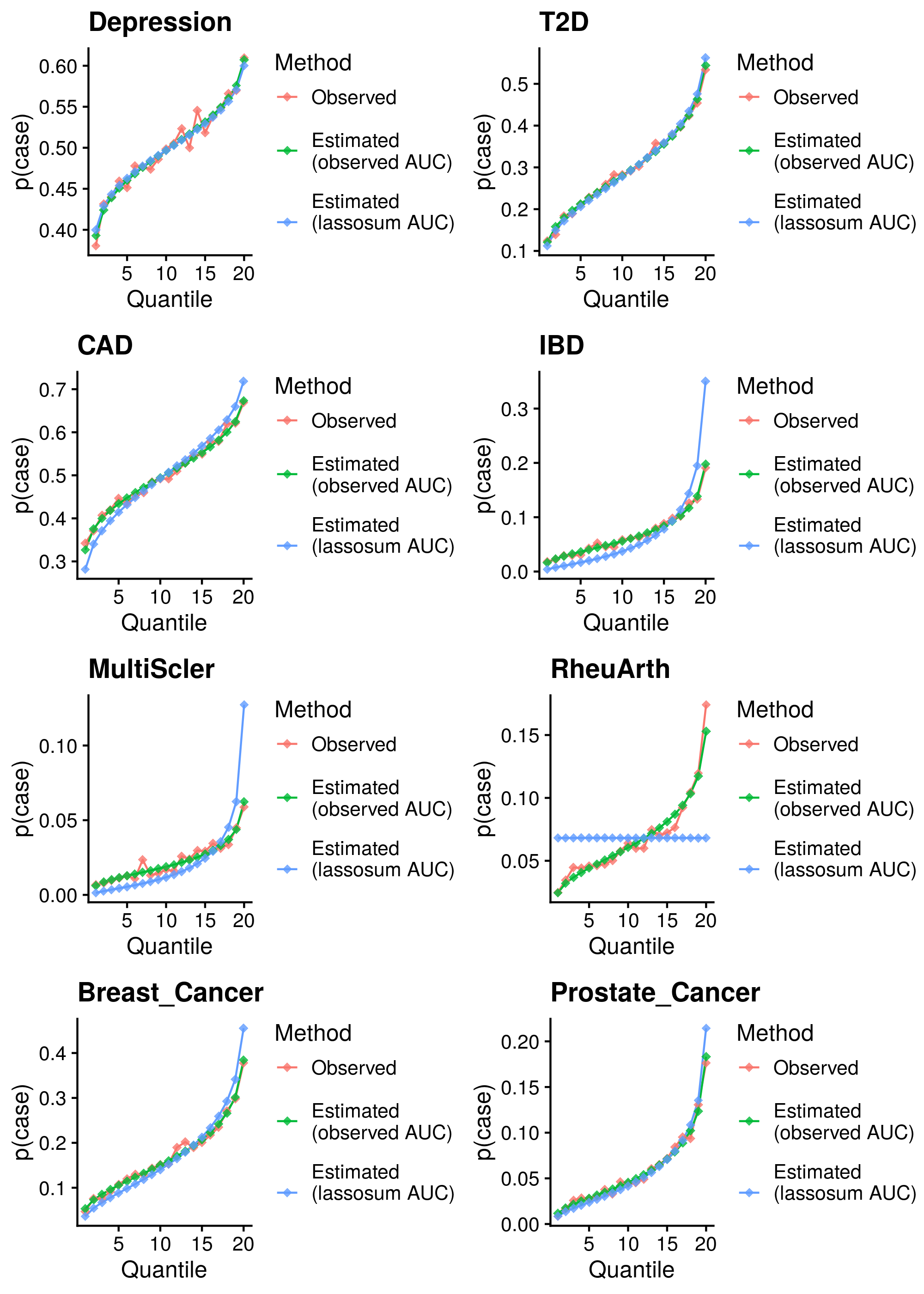
| Phenotype | Correlation (95%CI) | Mean Abs. Diff. | N | Ncas | Ncon |
|---|---|---|---|---|---|
| Depression | 0.985 (0.961-0.994) | 1.6% | 49999 | 24999 | 25000 |
| T2D | 0.996 (0.991-0.999) | 3.4% | 49999 | 14888 | 35111 |
| CAD | 0.995 (0.986-0.998) | 4.3% | 49999 | 25000 | 24999 |
| IBD | 0.963 (0.907-0.985) | 38.6% | 49999 | 3461 | 46538 |
| MultiScler | 0.915 (0.794-0.966) | 43.7% | 49999 | 1137 | 48862 |
| RheuArth | -0.008 (-0.449-0.436) | 40% | 49999 | 3408 | 46591 |
| Breast_Cancer | 0.991 (0.978-0.997) | 12.5% | 49999 | 8512 | 41487 |
| Prostate_Cancer | 0.99 (0.976-0.996) | 11.1% | 50000 | 2927 | 47073 |
Median Cor. = 0.9875522; Mean Cor. = 0.8534752; Min. Cor. = -0.007714644; Max. Cor. = 0.9964987
Excluding RheuArth: Median Cor. = 0.9904925; Mean Cor. = 0.9765024; Min. Cor. = 0.9150213; Max. Cor. = 0.9964987
2.1.5.2 Continuous
Show code
library(data.table)
library(e1071)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
gwas<-c('COLL01','HEIG03','BODY04')
pheno<-c('Intelligence','Height','BMI')
n_quant<-20
files<-data.frame(pheno,gwas)
# Create function
mean_sd_quant.f <- function(PRS_R2=0.641, Outcome_mean=1, Outcome_sd=1, n_quantile=20){
### PRS quantiles with a continuous phenotype (Y)
library(tmvtnorm)
###
E_PRS = 0
SD_PRS = sqrt(1)
E_phenotype = Outcome_mean
SD_phenotype = Outcome_sd
by_quant<-1/(n_quantile)
PRS_quantile_bounds <- qnorm(p=seq(0, 1, by=by_quant), mean= E_PRS, sd= SD_PRS)
lower_PRS_vec <- PRS_quantile_bounds[1:n_quantile]
upper_PRS_vec <- PRS_quantile_bounds[2:(n_quantile+1)]
mean_vec <- c(E_phenotype, E_PRS)
sigma_mat <- matrix(sqrt(PRS_R2)*SD_phenotype*SD_PRS, nrow=2, ncol=2)
sigma_mat[1,1] <- SD_phenotype^2
sigma_mat[2,2] <- SD_PRS^2
### mean of phenotype within the truncated PRS distribution
out_mean_Y <- rep(0, 20)
### SD of phenotype within the truncated PRS distribution
out_SD_Y <- rep(0, 20)
### cov of Y and PRS given truncation on PRS
out_cov_Y_PRS <- rep(0, 20)
### SD of PRS given truncation on PRS
out_SD_PRS <- rep(0, 20)
### mean PRS given truncation on PRS
out_mean_PRS <- rep(0, 20)
for(i in 1:n_quantile){
distribution_i <- mtmvnorm(mean = mean_vec,
sigma = sigma_mat,
lower = c(-Inf, lower_PRS_vec[i]),
upper = c(Inf, upper_PRS_vec[i]),
doComputeVariance=TRUE,
pmvnorm.algorithm=GenzBretz())
out_mean_Y[i] <- distribution_i$tmean[1]
out_mean_PRS[i] <- distribution_i$tmean[2]
out_SD_Y[i] <- sqrt(distribution_i$tvar[1,1])
out_SD_PRS[i] <- sqrt(distribution_i$tvar[2,2])
out_cov_Y_PRS[i] <- distribution_i$tvar[1,2]
}
out<-data.frame(q=1:n_quantile,
q_min=lower_PRS_vec,
q_max=upper_PRS_vec,
x_mean=out_mean_Y,
x_sd=out_SD_Y)
return(out)
out_mean_Y
out_SD_Y
out_mean_PRS
out_SD_PRS
out_cov_Y_PRS
}
# Read in lassosum R2
lassosum_R2<-read.csv('/scratch/users/k1806347/Analyses/AbsoluteRisk/Estimated_AUC_R2/lassosum/lassosum_R2.csv')
# Run analysis for each phenotype
res_all<-NULL
plots_all<-list()
cor_res<-NULL
for(i in 1:dim(files)[1]){
# Read in pheno and prs data, and merge
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',files$pheno[i],'.txt'))
names(pheno_i)[3]<-'pheno'
prs_i<-fread(paste0(UKBB_output,'/PRS_for_interpretation/1KG_ref/DBSLMM/',files$gwas[i],'/UKBB.subset.w_hm3.',files$gwas[i],'.DBSLMM_profiles'))
prs_i<-prs_i[,c('FID','IID',paste0(files$gwas[i], '_DBSLMM')), with=F]
names(prs_i)[3]<-'prs'
pheno_prs<-merge(pheno_i, prs_i, by=c('FID','IID'))
# Read in AUC for PRS
assoc<-fread(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/',files$pheno[i],'/UKBB.w_hm3.',files$gwas[i],'.EUR-PRSs.AllMethodComp.assoc.txt'))
prs_r2<-assoc[grepl('DBSLMM', assoc$Predictor),]$Obs_R2
# Assign individuals to observed PRS quantiles
obs_quant<-quantile(pheno_prs$prs, prob = seq(0, 1, length = n_quant+1))
pheno_prs$obs_quant<-as.numeric(cut( pheno_prs$prs, obs_quant, include.lowest = T))
# Calculate mean and SD of each quantile that are cases
obs_dist<-NULL
for(k in 1:n_quant){
obs_dist<-rbind(obs_dist, data.frame(Phenotype=files$pheno[i],
Type='Observed',
Quantile=k,
q_min=obs_quant[k],
q_max=obs_quant[k+1],
x_mean=mean(pheno_prs$pheno[pheno_prs$obs_quant == k]),
x_sd=sd(pheno_prs$pheno[pheno_prs$obs_quant == k])))
}
# Assign individuals to estimated PRS quantiles
est_dist<-mean_sd_quant.f(PRS_R2 = prs_r2, Outcome_mean=mean(pheno_prs$pheno), Outcome_sd=sd(pheno_prs$pheno), n_quantile = n_quant)
est_dist$q<-NULL
est_dist<-data.frame(Phenotype=files$pheno[i],Type='\nEstimated\n(Observed R2)\n',Quantile=1:n_quant, est_dist)
est_quant<-sort(unique(c(est_dist$q_min, est_dist$q_max)))
pheno_prs$est_quant<-as.numeric(cut( pheno_prs$prs, est_quant, include.lowest = T))
# Assign individuals to lassosum estimated PRS quantiles
lasso_est_dist<-mean_sd_quant.f(PRS_R2 = lassosum_R2$lasso_R2[lassosum_R2$Phenotype == files$pheno[i]], Outcome_mean=mean(pheno_prs$pheno), Outcome_sd=sd(pheno_prs$pheno), n_quantile = n_quant)
lasso_est_dist$q<-NULL
lasso_est_dist<-data.frame(Phenotype=files$pheno[i],Type="Estimated\n(lassosum R2)\n",Quantile=1:n_quant, lasso_est_dist)
lasso_est_quant<-sort(unique(c(lasso_est_dist$q_min, lasso_est_dist$q_max)))
pheno_prs$lasso_est_quant<-as.numeric(cut( pheno_prs$prs, lasso_est_quant, include.lowest = T))
quant_comp<-do.call(rbind, list(obs_dist, est_dist, lasso_est_dist))
tmp<-cor.test(obs_dist$x_mean, est_dist$x_mean)
tmp2<-cor.test(obs_dist$x_mean, lasso_est_dist$x_mean)
cor_res<-rbind(cor_res,data.frame(Phenotype=files$pheno[i],
Cor_mean=tmp$estimate,
Cor_mean_Low95CI=tmp$conf.int[1],
Cor_mean_High95CI=tmp$conf.int[2],
Mean_perc_diff_mean=mean(abs(est_dist$x_mean-obs_dist$x_mean)/obs_dist$x_mean),
Mean_perc_diff_sd=mean(abs(est_dist$x_sd-obs_dist$x_sd)/obs_dist$x_sd),
Cor_mean_lasso=tmp2$estimate,
Cor_mean_Low95CI_lasso=tmp2$conf.int[1],
Cor_mean_High95CI_lasso=tmp2$conf.int[2],
Mean_perc_diff_mean_lasso=mean(abs(lasso_est_dist$x_mean-obs_dist$x_mean)/obs_dist$x_mean),
Mean_perc_diff_sd_lasso=mean(abs(lasso_est_dist$x_sd-obs_dist$x_sd)/obs_dist$x_sd),
N=length(pheno_prs$pheno),
Skewness=skewness(pheno_prs$pheno)))
res_all<-rbind(res_all, quant_comp)
library(ggplot2)
library(cowplot)
plots_all[[i]]<-ggplot(quant_comp, aes(x=Quantile, y=x_mean, colour=Type)) +
geom_point(stat="identity", position=position_dodge(.5), alpha=0.8, shape=18, size=2) +
geom_errorbar(aes(ymin=x_mean-x_sd, ymax=x_mean+x_sd), width=.2, position=position_dodge(.5), alpha=0.8) +
labs(y="Mean (SD)", title=files$pheno[i], colour='Method') +
theme_cowplot(12)
}
png(paste0('/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/Mean_SD_Comp_lasso.png'), units='px', res=300, width=1750, height=2000)
plot_grid(plotlist=plots_all, ncol = 1)
dev.off()
write.csv(cor_res, '/scratch/users/k1806347/Analyses/AbsoluteRisk/Measured_AUC_R2/Mean_SD_Comp_lasso.csv', row.names=F, quote=F)Show results
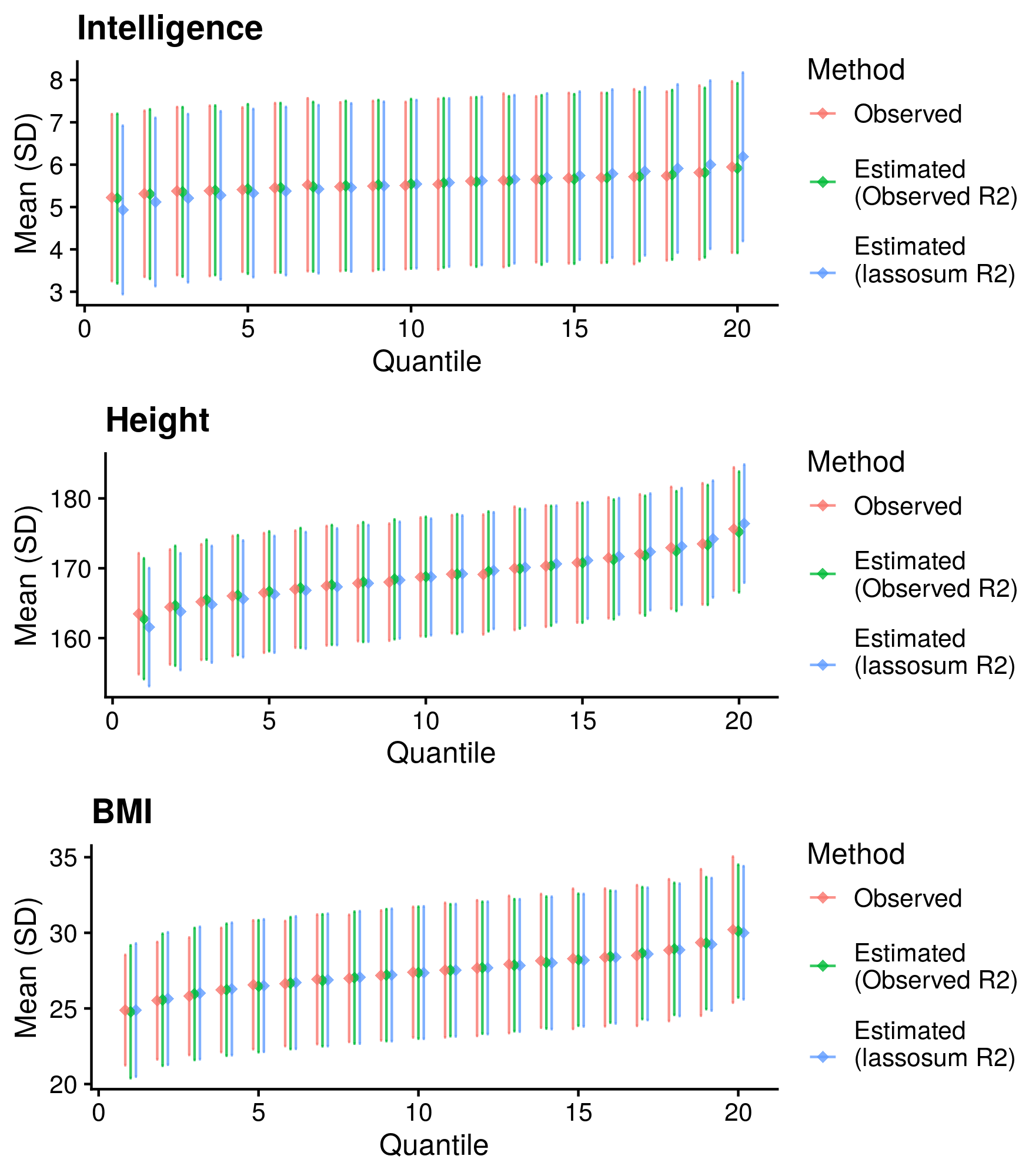
| Phenotype | Correlation (95%CI) | Mean Abs. Diff. of Mean | Mean Abs. Diff. of SD | N | Skewness |
|---|---|---|---|---|---|
| Intelligence | 0.992 (0.979-0.997) | 1.9% | 1.5% | 50000 | 0.144 |
| Height | 0.996 (0.989-0.998) | 0.2% | 2.5% | 49999 | 0.117 |
| BMI | 0.998 (0.995-0.999) | 0.3% | 6% | 49999 | 0.592 |
Median Cor. of means = 0.9958381; Mean Cor. of means = 0.9952452; Min. Cor. of means = 0.9917264; Max. Cor. of means = 0.9981713
## Warning in mean.default(res$PercDiff_sd_mean_lasso): argument is not numeric or
## logical: returning NA## Warning in min(res$PercDiff_sd_mean_lasso): no non-missing arguments to min;
## returning Inf## Warning in max(res$PercDiff_sd_mean_lasso): no non-missing arguments to max;
## returning -InfMedian mean %diff of SD = ; Mean mean %diff of SD = NA; Min. mean %diff of SD = Inf; Max. mean %diff of SD = -Inf
3 Prepare input for Impute.Me module
I need to download GWAS sumstats in an automated way, estimate the underlying heritability and produce reference standardised score files. GWAS sumstats will be downloaded from the GWAS catalogue.
3.1 Download GWAS sumstats
Show results
# Download latest GWAS catalogue index (22/11/2020)
# https://www.ebi.ac.uk/gwas/downloads/summary-statistics
# Read in the csv
gwas_list<-read.csv('~/brc_scratch/Data/GWAS_sumstats/GWAS_catalog/list_gwas_summary_statistics_22_Nov_2020.csv')
# Create a list GWAS catalogue IDs
gwas_ids<-c('GCST006901','GCST006900','GCST003045','GCST004773','GCST004296','GCST006572')
gwas_list_subset<-gwas_list[(gwas_list$Study.accession %in% gwas_ids),]
# List ftp downloads page
system(paste0('wget ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/ -P ~/brc_scratch/Data/GWAS_sumstats/GWAS_catalog/'))
ftp_list<-read.table('~/brc_scratch/Data/GWAS_sumstats/GWAS_catalog/index.html', sep='&', header=F)$V1
ftp_list<-ftp_list[grepl(paste(gwas_list_subset$Study.accession, collapse='|'), ftp_list)]
ftp_list<-gsub('>.*','',gsub('.*href=','',ftp_list))
# Download these summary statistics
for(gwas_i in ftp_list){
system(paste0('wget ',gwas_i,' -O ~/brc_scratch/Data/GWAS_sumstats/GWAS_catalog/temp.txt'))
temp_list<-read.table('~/brc_scratch/Data/GWAS_sumstats/GWAS_catalog/temp.txt', sep='&', header=F)$V1
temp_list<-temp_list[grepl('href', temp_list)]
temp_list<-gsub('>.*','',gsub('.*href=','',temp_list))
if(sum(grepl('harmonised', temp_list) == T) > 0){
system(paste0('wget -r ',gwas_i,'harmonised/ -P ~/brc_scratch/Data/GWAS_sumstats/GWAS_catalog/'))
} else {
system(paste0('wget -r ',gwas_i,' -P ~/brc_scratch/Data/GWAS_sumstats/GWAS_catalog/'))
}
}
# After looking through the results for these studies, they are often not full summary statistics. Some are not in harmonised format, and some contain trans ancestry data. I think using the KCL repository might be a better option.